 |
PDBsum entry 1qiq

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Antibiotic biosynthesis
|
PDB id
|
|

|

|
1qiq
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.21.3.1
- isopenicillin-N synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine + O2 = isopenicillin N + 2 H2O
|
 |
 |
 |
 |
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine
|
+
|
 O2
O2
|
=
|
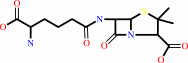 isopenicillin N
isopenicillin N
|
+
|
2
×
H2O
Bound ligand (Het Group name = )
matches with 84.62% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
401:721-724
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
The reaction cycle of isopenicillin N synthase observed by X-ray diffraction.
|
|
N.I.Burzlaff,
P.J.Rutledge,
I.J.Clifton,
C.M.Hensgens,
M.Pickford,
R.M.Adlington,
P.L.Roach,
J.E.Baldwin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Isopenicillin N synthase (IPNS), a non-haem iron-dependent oxidase, catalyses
the biosynthesis of isopenicillin N (IPN), the precursor of all penicillins and
cephalosporins. The key steps in this reaction are the two
iron-dioxygen-mediated ring closures of the tripeptide
delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-valine (ACV). It has been proposed
that the four-membered beta-lactam ring forms initially, associated with a
highly oxidized iron(iv)-oxo (ferryl) moiety, which subsequently mediates
closure of the five-membered thiazolidine ring. Here we describe observation of
the IPNS reaction in crystals by X-ray crystallography. IPNS Fe2+ substrate
crystals were grown anaerobically, exposed to high pressures of oxygen to
promote reaction and frozen, and their structures were elucidated by X-ray
diffraction. Using the natural substrate ACV, this resulted in the IPNS x Fe2+ x
IPN product complex. With the substrate analogue,
delta-(L-alpha-aminoadipoyl)-L-cysteinyl-L-S-methylcysteine (ACmC) in the
crystal, the reaction cycle was interrupted at the monocyclic stage. These mono-
and bicyclic structures support our hypothesis of a two-stage reaction sequence
leading to penicillin. Furthermore, the formation of a monocyclic sulphoxide
product from ACmC is most simply explained by the interception of a high-valency
iron-oxo species.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 Proposed mechanisms for the oxidation of ACV and
ACmC to bicyclic and monocyclic products, respectively. See text
for details of compounds 1-6. AA, L-  -( -(
 -aminoadipoyl). -aminoadipoyl).
|
 |
Figure 3.
Figure 3 Stereo views of the two substrates and two products
overlaid. The key regions that participate in the reaction and
the iron atom (orange) are shown; the aminoadipoyl side chain,
which does not move significantly, is omitted for clarity. Shown
are ACV (white), IPN (yellow), ACmC (blue) and its monocyclic
sulphoxide product (pink). Figures were prepared using the
programs MOLSCRIPT20 and Raster3D (ref. 21).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(1999,
401,
721-724)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.K.Sydor,
S.M.Barry,
O.M.Odulate,
F.Barona-Gomez,
S.W.Haynes,
C.Corre,
L.Song,
and
G.L.Challis
(2011).
Regio- and stereodivergent antibiotic oxidative carbocyclizations catalysed by Rieske oxygenase-like enzymes.
|
| |
Nat Chem,
3,
388-392.
|
 |
|
|
|
|
 |
W.A.Schenk
(2011).
The coordination chemistry of small sulfur-containing molecules: a personal perspective.
|
| |
Dalton Trans,
40,
1209-1219.
|
 |
|
|
|
|
 |
A.Benjdia,
S.Subramanian,
J.Leprince,
H.Vaudry,
M.K.Johnson,
and
O.Berteau
(2010).
Anaerobic sulfatase-maturating enzyme--a mechanistic link with glycyl radical-activating enzymes?
|
| |
FEBS J,
277,
1906-1920.
|
 |
|
|
|
|
 |
C.Yi,
G.Jia,
G.Hou,
Q.Dai,
W.Zhang,
G.Zheng,
X.Jian,
C.G.Yang,
Q.Cui,
and
C.He
(2010).
Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
|
| |
Nature,
468,
330-333.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.J.Dungan,
Y.Ortin,
H.Mueller-Bunz,
and
P.J.Rutledge
(2010).
Design and synthesis of a tetradentate '3-amine-1-carboxylate' ligand to mimic the metal binding environment at the non-heme iron(II) oxidase active site.
|
| |
Org Biomol Chem,
8,
1666-1673.
|
 |
|
|
|
|
 |
B.Sjöblom,
M.Polentarutti,
and
K.Djinovic-Carugo
(2009).
Structural study of X-ray induced activation of carbonic anhydrase.
|
| |
Proc Natl Acad Sci U S A,
106,
10609-10613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Morokuma
(2009).
Theoretical studies of structure, function and reactivity of molecules-A personal account.
|
| |
Proc Jpn Acad Ser B Phys Biol Sci,
85,
167-182.
|
 |
|
|
|
|
 |
R.M.Cicchillo,
H.Zhang,
J.A.Blodgett,
J.T.Whitteck,
G.Li,
S.K.Nair,
W.A.van der Donk,
and
W.W.Metcalf
(2009).
An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis.
|
| |
Nature,
459,
871-874.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Ge,
I.J.Clifton,
A.R.Howard-Jones,
J.E.Stok,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2009).
Structural studies on the reaction of isopenicillin N synthase with a sterically demanding depsipeptide substrate analogue.
|
| |
Chembiochem,
10,
2025-2031.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.W.Metcalf,
and
W.A.van der Donk
(2009).
Biosynthesis of phosphonic and phosphinic acid natural products.
|
| |
Annu Rev Biochem,
78,
65-94.
|
 |
|
|
|
|
 |
A.Siddiq,
L.R.Aminova,
and
R.R.Ratan
(2008).
Prolyl 4-hydroxylase activity-responsive transcription factors: from hydroxylation to gene expression and neuroprotection.
|
| |
Front Biosci,
13,
2875-2887.
|
 |
|
|
|
|
 |
K.Watanabe
(2008).
Exploring the biosynthesis of natural products and their inherent suitability for the rational design of desirable compounds through genetic engineering.
|
| |
Biosci Biotechnol Biochem,
72,
2491-2506.
|
 |
|
|
|
|
 |
L.Gidijala,
R.A.Bovenberg,
P.Klaassen,
I.J.van der Klei,
M.Veenhuis,
and
J.A.Kiel
(2008).
Production of functionally active Penicillium chrysogenum isopenicillin N synthase in the yeast Hansenula polymorpha.
|
| |
BMC Biotechnol,
8,
29.
|
 |
|
|
|
|
 |
P.C.Bruijnincx,
G.van Koten,
and
R.J.Klein Gebbink
(2008).
Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies.
|
| |
Chem Soc Rev,
37,
2716-2744.
|
 |
|
|
|
|
 |
A.C.Stewart,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2007).
A cyclobutanone analogue mimics penicillin in binding to isopenicillin N synthase.
|
| |
Chembiochem,
8,
2003-2007.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Ozer,
and
R.K.Bruick
(2007).
Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one?
|
| |
Nat Chem Biol,
3,
144-153.
|
 |
|
|
|
|
 |
D.Hoffmeister,
and
N.P.Keller
(2007).
Natural products of filamentous fungi: enzymes, genes, and their regulation.
|
| |
Nat Prod Rep,
24,
393-416.
|
 |
|
|
|
|
 |
D.P.Galonić,
E.W.Barr,
C.T.Walsh,
J.M.Bollinger,
and
C.Krebs
(2007).
Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3.
|
| |
Nat Chem Biol,
3,
113-116.
|
 |
|
|
|
|
 |
A.Daruzzaman,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2006).
Unexpected oxidation of a depsipeptide substrate analogue in crystalline isopenicillin N synthase.
|
| |
Chembiochem,
7,
351-358.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.J.Kershaw,
M.E.Caines,
M.C.Sleeman,
and
C.J.Schofield
(2005).
The enzymology of clavam and carbapenem biosynthesis.
|
| |
Chem Commun (Camb),
(),
4251-4263.
|
 |
|
|
|
|
 |
K.Valegård,
A.C.Terwisscha van Scheltinga,
A.Dubus,
G.Ranghino,
L.M.Oster,
J.Hajdu,
and
I.Andersson
(2004).
The structural basis of cephalosporin formation in a mononuclear ferrous enzyme.
|
| |
Nat Struct Mol Biol,
11,
95.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.J.Clifton,
L.X.Doan,
M.C.Sleeman,
M.Topf,
H.Suzuki,
R.C.Wilmouth,
and
C.J.Schofield
(2003).
Crystal structure of carbapenem synthase (CarC).
|
| |
J Biol Chem,
278,
20843-20850.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.M.Hensgens,
E.A.Kroezinga,
B.A.van Montfort,
J.M.van der Laan,
J.D.Sutherland,
and
B.W.Dijkstra
(2002).
Purification, crystallization and preliminary X-ray diffraction of Cys103Ala acyl coenzyme A: isopenicillin N acyltransferase from Penicillium chrysogenum.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
716-718.
|
 |
|
|
|
|
 |
M.J.Ryle,
and
R.P.Hausinger
(2002).
Non-heme iron oxygenases.
|
| |
Curr Opin Chem Biol,
6,
193-201.
|
 |
|
|
|
|
 |
A.M.Cerdeño,
M.J.Bibb,
and
G.L.Challis
(2001).
Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes.
|
| |
Chem Biol,
8,
817-829.
|
 |
|
|
|
|
 |
J.M.Ogle,
I.J.Clifton,
P.J.Rutledge,
J.M.Elkins,
N.I.Burzlaff,
R.M.Adlington,
P.L.Roach,
and
J.E.Baldwin
(2001).
Alternative oxidation by isopenicillin N synthase observed by X-ray diffraction.
|
| |
Chem Biol,
8,
1231-1237.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.O.Bachmann,
and
C.A.Townsend
(2000).
Kinetic mechanism of the beta-lactam synthetase of Streptomyces clavuligerus.
|
| |
Biochemistry,
39,
11187-11193.
|
 |
|
|
|
|
 |
G.A.Petsko,
and
D.Ringe
(2000).
Observation of unstable species in enzyme-catalyzed transformations using protein crystallography.
|
| |
Curr Opin Chem Biol,
4,
89-94.
|
 |
|
|
|
|
 |
I.Schlichting,
and
K.Chu
(2000).
Trapping intermediates in the crystal: ligand binding to myoglobin.
|
| |
Curr Opin Struct Biol,
10,
744-752.
|
 |
|
|
|
|
 |
W.A.Schenk
(2000).
Isopenicillin N Synthase: An Enzyme at Work.
|
| |
Angew Chem Int Ed Engl,
39,
3409-3411.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

