 |
PDBsum entry 1oc1

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1oc1
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.21.3.1
- isopenicillin-N synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine + O2 = isopenicillin N + 2 H2O
|
 |
 |
 |
 |
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine
|
+
|
 O2
O2
|
=
|
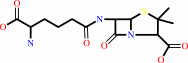 isopenicillin N
isopenicillin N
|
+
|
2
×
H2O
Bound ligand (Het Group name = )
matches with 95.83% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem J
372:687-693
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies on the reaction of isopenicillin N synthase with the substrate analogue delta-(l-alpha-aminoadipoyl)-l-cysteinyl-d-alpha-aminobutyrate.
|
|
A.J.Long,
I.J.Clifton,
P.L.Roach,
J.E.Baldwin,
C.J.Schofield,
P.J.Rutledge.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Isopenicillin N synthase (IPNS) is a non-haem iron(II) oxidase which catalyses
the biosynthesis of isopenicillin N from the tripeptide
delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-valine (ACV). Herein we report
crystallographic studies to investigate the reaction of IPNS with the truncated
substrate analogue
delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-alpha-aminobutyrate (ACAb). It has
been reported previously that this analogue gives rise to three beta-lactam
products when incubated with IPNS: two methyl penams and a cepham. Crystal
structures of the IPNS-Fe(II)-ACAb and IPNS-Fe(II)-ACAb-NO complexes have now
been solved and are reported herein. These structures and modelling studies
based on them shed light on the diminished product selectivity shown by IPNS in
its reaction with ACAb and further rationalize the presence of certain key
residues at the IPNS active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.D.Brown-Marshall,
A.R.Diebold,
and
E.I.Solomon
(2010).
Reaction coordinate of isopenicillin N synthase: oxidase versus oxygenase activity.
|
| |
Biochemistry,
49,
1176-1182.
|
 |
|
|
|
|
 |
W.Ge,
I.J.Clifton,
A.R.Howard-Jones,
J.E.Stok,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2009).
Structural studies on the reaction of isopenicillin N synthase with a sterically demanding depsipeptide substrate analogue.
|
| |
Chembiochem,
10,
2025-2031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.C.Stewart,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2007).
A cyclobutanone analogue mimics penicillin in binding to isopenicillin N synthase.
|
| |
Chembiochem,
8,
2003-2007.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Daruzzaman,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2006).
Unexpected oxidation of a depsipeptide substrate analogue in crystalline isopenicillin N synthase.
|
| |
Chembiochem,
7,
351-358.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

