
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
First structural evidence of a specific inhibition of phospholipase a2 by alpha-Tocopherol (vitamin e) and its implications in inflammation: crystal structure of the complex formed between phospholipase a2 and alpha-Tocopherol at 1.8 a resolution.
|
 |
|
Authors
|
 |
V.Chandra,
J.Jasti,
P.Kaur,
C.H.Betzel,
A.Srinivasan,
T.P.Singh.
|
 |
|
Ref.
|
 |
J Mol Biol, 2002,
320,
215-222.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
This is the first structural evidence of alpha-tocopherol (alpha-TP) as a
possible candidate against inflammation, as it inhibits phospholipase A2
specifically and effectively. The crystal structure of the complex formed
between Vipera russelli phospholipase A2 and alpha-tocopherol has been
determined and refined to a resolution of 1.8 A. The structure contains two
molecules, A and B, of phospholipase A2 in the asymmetric unit, together with
one alpha-tocopherol molecule, which is bound specifically to one of them. The
phospholipase A2 molecules interact extensively with each other in the
crystalline state. The two molecules were found in a stable association in the
solution state as well, thus indicating their inherent tendency to remain
together as a structural unit, leading to significant functional implications.
In the crystal structure, the most important difference between the
conformations of two molecules as a result of their association pertains to the
orientation of Trp31. It may be noted that Trp31 is located at the mouth of the
hydrophobic channel that forms the binding domain of the enzyme. The values of
torsion angles (phi, psi, chi(1) and chi(2)) for both the backbone as well as
for the side-chain of Trp31 in molecules A and B are -94 degrees, -30 degrees,
-66 degrees, 116 degrees and -128 degrees, 170 degrees, -63 degrees, -81
degrees, respectively. The conformation of Trp31 in molecule A is suitable for
binding, while that in B hinders the passage of the ligand to the binding site.
Consequently, alpha-tocopherol is able to bind to molecule A only, while the
binding site of molecule B contains three water molecules. In the complex, the
aromatic moiety of alpha-tocopherol is placed in the large space at the active
site of the enzyme, while the long hydrophobic channel in the enzyme is filled
by hydrocarbon chain of alpha-tocopherol. The critical interactions between the
enzyme and alpha-tocopherol are generated between the hydroxyl group of the
six-membered ring of alpha-tocopherol and His48 N(delta1) and Asp49 O(delta1) as
characteristic hydrogen bonds. The remaining part of alpha-tocopherol interacts
extensively with the residues of the hydrophobic channel of the enzyme, giving
rise to a number of hydrophobic interactions, resulting in the formation of a
stable complex.
|
 |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. (F[o] -F[c]) electron density map contoured at
2.5s showing water molecules in the binding region of molecule
B. The Figure was drawn with BOBSCRIPT21 and rendered by
Raster3D.22
|
 |
Figure 6.
Figure 6. The binding of a-tocopherol in molecule A. The OH
group of the aromatic moiety in a-tocopherol plays a key role by
interacting simultaneously with both Asp49 and His48. The
hydrocarbon chain of a-tocopherol fills the hydrophobic channel
in the protein. The Figure was drawn with MOLSCRIPT23 and
rendered by Raster3D.22
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
320,
215-222)
copyright 2002.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
First structural evidence of antiinflammatory action of vitamin e (2,5,7,8-Tetramethyl-2-(4',8',12'- Trimethyltridecyl)-6-Chromanol) through its binding to phospholipase a2 specifically: crystal structure of a complex formed between phospholipase a2 and vitamin e at 1.80 resolution
|
 |
|
Authors
|
 |
V.Chandra,
J.Jasti,
P.Kaur,
C.Betzel,
A.Srinivasan,
T.P.Singh.
|
 |
|
Ref.
|
 |
TO BE PUBLISHED ...
|
 |
 |
|
Secondary reference #2
|
 |
|
Title
|
 |
Crystal structure of a complex formed between phospholipase a2 from daboia russelli pulchella and a designed pentapeptide phe-Leu-Ser-Tyr-Lys at 1.8 resolution
|
 |
|
Authors
|
 |
V.Chandra,
J.Jasti,
P.Kaur,
C.Betzel,
A.Srinivasan,
T.P.Singh.
|
 |
|
Ref.
|
 |
TO BE PUBLISHED ...
|
 |
 |
|
Secondary reference #3
|
 |
|
Title
|
 |
First structural evidence of the inhibition of phospholipase a2 by aristolochic acid: crystal structure of a complex formed between phospholipase a2 and aristolochic acid
|
 |
|
Authors
|
 |
V.Chandra,
J.Jasti,
P.Kaur,
C.Betzel,
A.Srinivasan,
T.P.Singh.
|
 |
|
Ref.
|
 |
TO BE PUBLISHED ...
|
 |
 |
|
Secondary reference #4
|
 |
|
Title
|
 |
Design of specific peptide inhibitors of phospholipase a2: structure of a complex formed between russell'S viper phospholipase a2 and a designed peptide leu-Ala-Ile-Tyr-Ser (laiys).
|
 |
|
Authors
|
 |
V.Chandra,
J.Jasti,
P.Kaur,
S.Dey,
A.Srinivasan,
C.H.Betzel,
T.P.Singh.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2002,
58,
1813-1819.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
|
Note In the PDB file this reference is
annotated as "TO BE PUBLISHED".
The citation details given above were identified by an automated
search of PubMed on title and author
names, giving a
percentage match of
71%.
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 The (F[o] - F[c]) map contoured at 2.5  showing
the electron density for the peptide LAIYS in molecule A. showing
the electron density for the peptide LAIYS in molecule A.
|
 |
Figure 4.
Figure 4 (a) Interactions between DPLA[2] and the designed
peptide LAIYS. The peptide residues are indicated with a `P' in
parentheses. The critical interactions between Tyr(P) OH and
His N 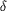 1
and Asp O 1
and Asp O 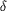 1,
including other hydrogen bonds between peptide and protein, are
indicated by dotted lines. The figure was drawn with MOLSCRIPT
(Kraulis, 1991[Kraulis, P. J. (1991). J. Appl. Cryst. 24,
946-950.]) and Raster3D (Merritt & Murphy, 1994[Merritt, E. A. &
Murphy, M. E. P. (1994). Acta Cryst. D50, 869-873.]). (b)
LIGPLOT (Wallace et al., 1995[Wallace, A. C., Laskowski, R. A. &
Thornton, J. M. (1995). Protein Eng. 8, 127-134.]) showing the
schematic representation of the interactions between the peptide
LAIYS and protein molecule. (c) GRASP (Nicholls et al.,
1991[Nicholls, A., Sharp, K. & Honig, B. (1991). Proteins, 11,
281-296.]) representation of the binding cavity and the
hydrophobic channel. The peptide LAIYS is almost completely
buried in the pocket. Two key hydrogen bonds involving His48 and
Asp49 of the protein with peptide Tyr OH are also indicated by
dotted lines. 1,
including other hydrogen bonds between peptide and protein, are
indicated by dotted lines. The figure was drawn with MOLSCRIPT
(Kraulis, 1991[Kraulis, P. J. (1991). J. Appl. Cryst. 24,
946-950.]) and Raster3D (Merritt & Murphy, 1994[Merritt, E. A. &
Murphy, M. E. P. (1994). Acta Cryst. D50, 869-873.]). (b)
LIGPLOT (Wallace et al., 1995[Wallace, A. C., Laskowski, R. A. &
Thornton, J. M. (1995). Protein Eng. 8, 127-134.]) showing the
schematic representation of the interactions between the peptide
LAIYS and protein molecule. (c) GRASP (Nicholls et al.,
1991[Nicholls, A., Sharp, K. & Honig, B. (1991). Proteins, 11,
281-296.]) representation of the binding cavity and the
hydrophobic channel. The peptide LAIYS is almost completely
buried in the pocket. Two key hydrogen bonds involving His48 and
Asp49 of the protein with peptide Tyr OH are also indicated by
dotted lines.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|
Secondary reference #5
|
 |
|
Title
|
 |
Regulation of catalytic function by molecular association: structure of phospholipase a2 from daboia russelli pulchella (dpla2) at 1.9 a resolution.
|
 |
|
Authors
|
 |
V.Chandra,
P.Kaur,
J.Jasti,
C.Betzel,
T.P.Singh.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2001,
57,
1793-1798.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 A region of the final 2F[o] - F[c] electron-density map
contoured at 1.5  and
the corresponding refined model. The diagram was produced using
the program O (Jones et al., 1991[Jones, T. A., Zou, J., Cowan,
S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110-119.]). and
the corresponding refined model. The diagram was produced using
the program O (Jones et al., 1991[Jones, T. A., Zou, J., Cowan,
S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110-119.]).
|
 |
Figure 8.
Figure 8 Positioning of Trp31 vis-à-vis the hydrophobic binding
channel. The placement of Trp31 in molecule B (blue) reduces the
width of the channel, thus impairing its binding capability
while the corresponding width in molecule A (green) is optimum.
The distances between the two nearest atoms of Leu2 and Trp31
are 8.3 and 4.7 Å in molecules A and B, respectively.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|
Secondary reference #6
|
 |
|
Title
|
 |
Three-Dimensional structure of a presynaptic neurotoxic phospholipase a2 from daboia russelli pulchella at 2.4 a resolution.
|
 |
|
Authors
|
 |
V.Chandra,
P.Kaur,
A.Srinivasan,
T.P.Singh.
|
 |
|
Ref.
|
 |
J Mol Biol, 2000,
296,
1117-1126.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. (a) Superimposition of C^α traces of DPLA[2]
(thick lines) and VPLA[2] (thin lines). The r.m.s. difference
for the C^α atoms is 1.2 Å. The corresponding shifts for
the neurotoxic (55–61 and 85–94), and anticoagulant
(53–77) fragments are 1.8 Å and 1.3 Å,
respectively. (b) Protruding side-chains of basic residues for
the anticoagulant site.
|
 |
Figure 4.
Figure 4. (a) The association of molecules A and B showing a
number of residues from both the molecules (black) and solvent
molecules (red) involved in the interactions between two
molecules: molecule A, Leu2, Leu17, Ala18, Ile19, Pro20, Trp31,
Arg43, Phe46, Ser70, Arg72, Met118, Leu119 and Asp122; molecule
B, Thr36, Ala40, Arg43, Phe46, Val47, Asn54, Glu97, Lys100,
Ile104, Gln108, Asn111, Leu130, Lys131 and Cys133 and 31 solvent
molecules. (b) Spatially two adjacent fragments 55–61 and
85–94. The segment 55–61 forms a β-turn I with a hydrogen
bond between Leu55 (O) and Cys61 (N). A tight loop, 85-94, is
stabilized by a number of intra-loop hydrogen bonds which are
indicated by dotted lines. The inter-segmental hydrogen bonds
(red, broken lines) are also indicated.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from Elsevier
|
 |
|

|
|
|

