 |
PDBsum entry 1ib4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.15
- endo-polygalacturonase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1,4-alpha-D-galacturonosyl)n+m + H2O = (1,4-alpha-D-galacturonosyl)n + (1,4-alpha-D-galacturonosyl)m
|
 |
 |
 |
 |
 |
(1,4-alpha-D-galacturonosyl)n+m
|
+
|
H2O
|
=
|
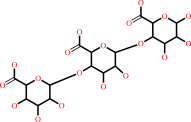 (1,4-alpha-D-galacturonosyl)n
(1,4-alpha-D-galacturonosyl)n
|
+
|
(1,4-alpha-D-galacturonosyl)m
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
311:863-878
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
The X-ray structure of Aspergillus aculeatus polygalacturonase and a modeled structure of the polygalacturonase-octagalacturonate complex.
|
|
S.W.Cho,
S.Lee,
W.Shin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Polygalacturonases hydrolyze the alpha-(1-4) glycosidic bonds of de-esterified
pectate in the smooth region of the plant cell wall. Crystal structures of
polygalacturonase from Aspergillus aculeatus were determined at pH 4.5 and 8.5
both to 2.0 A resolution. A. aculeatus polygalacturonase is a glycoprotein with
one N and ten O-glycosylation sites and folds into a right-handed parallel
beta-helix. The structures of the three independent molecules are essentially
the same, showing no dependency on pH or crystal packing, and are very similar
to that of Aspergillus niger polygalacturonase. However, the structures of the
long T1 loop containing a catalytic tyrosine residue are significantly different
in the two proteins. A three-dimensional model showing the substrate binding
mode for a family 28 hydrolase was obtained by a combined approach of flexible
docking, molecular dynamics simulations, and energy minimization. The
octagalacturonate substrate was modeled as an unbent irregular helix with the -1
ring in a half-chair ((4)H(3)) form that approaches the transition state
conformation. A comparative modeling of the three polygalacturonases with known
structure shows that six subsites ranging from -4 to +2 are clearly defined but
subsites -5 and +3 may or may not be shaped depending on the nearby amino acid
residues. Both distal subsites are mostly exposed to the solvent region and have
weak binding affinity even if they exist. The complex model provides a clear
explanation for the functions, either in catalysis or in substrate binding, of
all conserved amino acid residues in the polygalacturonase family of proteins.
Modeling suggests that the role of the conserved Asn157 and Tyr270, which had
previously been unidentified, may be in transition state stabilization. In A.
niger polygalacturonase, the long T1 loop may have to undergo conformational
change upon binding of the substrate to bring the tyrosine residue close to
subsite -1.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. (a) A ribbon diagram of the Aspergillus aculeatus
polygalacturonase structure viewed onto b-sheet PB1. (b) Stereo
view showing the cross-section of the b-helix and the aligned
residues viewed from the N-terminal side. Four complete turns in
the middle of the b-helix are shown with the labels of b-sheets
and turns.
|
 |
Figure 3.
Figure 3. Stereo view of a modeled structure of the
PGA-octagalacturonate complex. The electrostatic potential is
drawn at the solvent accessible surface of polygalacturonase
from -9kT/e^ - (red) to +9kT/e^ - (blue) and the substrate is
represented with a space-filling model. The N terminus is on the
top and the C terminus on the bottom. The unbent substrate spans
the binding cleft that is formed by the protruding loop regions
T1 (left side) and T4 (right side). The Figure was produced with
GRASP.[57]
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2001,
311,
863-878)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.A.Mertens,
and
M.J.Bowman
(2011).
Expression and characterization of fifteen Rhizopus oryzae 99-880 polygalacturonase enzymes in Pichia pastoris.
|
| |
Curr Microbiol,
62,
1173-1178.
|
 |
|
|
|
|
 |
S.Aminzadeh,
H.Naderi-Manesh,
K.Khajeh,
B.Ranjbar,
and
N.Farrokhi
(2010).
Characterization of acid-induced partially folded conformation resembling a molten globule state of polygalacturonase from a filamentous fungus Tetracoccosporium sp.
|
| |
Appl Biochem Biotechnol,
160,
1921-1932.
|
 |
|
|
|
|
 |
N.Killiny,
and
R.P.Almeida
(2009).
Host structural carbohydrate induces vector transmission of a bacterial plant pathogen.
|
| |
Proc Natl Acad Sci U S A,
106,
22416-22420.
|
 |
|
|
|
|
 |
D.W.Abbott,
and
A.B.Boraston
(2008).
Structural biology of pectin degradation by Enterobacteriaceae.
|
| |
Microbiol Mol Biol Rev,
72,
301.
|
 |
|
|
|
|
 |
H.Trigui-Lahiani,
M.Ayadi,
N.Hadj-Taïeb,
M.B.Ali,
and
A.Gargouri
(2008).
Genomic organization of a polygalacturonase gene from a hyperpectinolytic mutant strain of Penicillium occitanis.
|
| |
FEMS Microbiol Lett,
281,
23-29.
|
 |
|
|
|
|
 |
P.B.Vordtriede,
and
M.D.Yoder
(2008).
Crystallization, X-ray diffraction analysis and preliminary structure determination of the polygalacturonase PehA from Agrobacterium vitis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
645-647.
|
 |
|
|
|
|
 |
L.Federici,
A.Di Matteo,
J.Fernandez-Recio,
D.Tsernoglou,
and
F.Cervone
(2006).
Polygalacturonase inhibiting proteins: players in plant innate immunity?
|
| |
Trends Plant Sci,
11,
65-70.
|
 |
|
|
|
|
 |
L.D.Kluskens,
G.J.van Alebeek,
J.Walther,
A.G.Voragen,
W.M.de Vos,
and
J.van der Oost
(2005).
Characterization and mode of action of an exopolygalacturonase from the hyperthermophilic bacterium Thermotoga maritima.
|
| |
FEBS J,
272,
5464-5473.
|
 |
|
|
|
|
 |
S.A.Douthit,
M.Dlakic,
D.E.Ohman,
and
M.J.Franklin
(2005).
Epimerase active domain of Pseudomonas aeruginosa AlgG, a protein that contains a right-handed beta-helix.
|
| |
J Bacteriol,
187,
4573-4583.
|
 |
|
|
|
|
 |
F.Alberto,
C.Bignon,
G.Sulzenbacher,
B.Henrissat,
and
M.Czjzek
(2004).
The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases.
|
| |
J Biol Chem,
279,
18903-18910.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.K.Choi,
B.H.Lee,
C.H.Chae,
and
W.Shin
(2004).
Computer modeling of the rhamnogalacturonase-"hairy" pectin complex.
|
| |
Proteins,
55,
22-33.
|
 |
|
|
|
|
 |
M.A.McDonough,
C.Ryttersgaard,
M.E.Bjørnvad,
L.Lo Leggio,
M.Schülein,
S.O.Schrøder Glad,
and
S.Larsen
(2002).
Crystallization and preliminary X-ray characterization of a thermostable pectate lyase from Thermotoga maritima.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
709-711.
|
 |
|
|
|
|
 |
T.Shimizu,
T.Nakatsu,
K.Miyairi,
T.Okuno,
and
H.Kato
(2002).
Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
|
| |
Biochemistry,
41,
6651-6659.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

