 |
PDBsum entry 1hqh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.3.1
- arginase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arginine + H2O = urea + L-ornithine
|
 |
 |
 |
 |
 |
 L-arginine
L-arginine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
=
|
 urea
urea
|
+
|
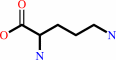 L-ornithine
L-ornithine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
40:2689-2701
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
|
|
J.D.Cox,
E.Cama,
D.M.Colleluori,
S.Pethe,
J.L.Boucher,
D.Mansuy,
D.E.Ash,
D.W.Christianson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Arginase is a binuclear Mn(2+) metalloenzyme that catalyzes the hydrolysis of
L-arginine to L-ornithine and urea. X-ray crystal structures of arginase
complexed to substrate analogues N(omega)-hydroxy-L-arginine and
N(omega)-hydroxy-nor-L-arginine, as well as the products L-ornithine and urea,
complete a set of structural "snapshots" along the reaction coordinate of
arginase catalysis when interpreted along with the X-ray crystal structure of
the arginase-transition-state analogue complex described in Kim et al. [Kim, N.
N., Cox, J. D., Baggio, R. F., Emig, F. A., Mistry, S., Harper, S. L., Speicher,
D. W., Morris, Jr., S. M., Ash, D. E., Traish, A. M., and Christianson, D. W.
(2001) Biochemistry 40, 2678-2688]. Taken together, these structures render
important insight on the structural determinants of tight binding inhibitors.
Furthermore, we demonstrate for the first time the structural mechanistic link
between arginase and NO synthase through their respective complexes with
N(omega)-hydroxy-L-arginine. That N(omega)-hydroxy-L-arginine is a catalytic
intermediate for NO synthase and an inhibitor of arginase reflects the
reciprocal metabolic relationship between these two critical enzymes of
L-arginine catabolism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Riley,
S.C.Roberts,
and
B.Ullman
(2011).
Inhibition profile of Leishmania mexicana arginase reveals differences with human arginase I.
|
| |
Int J Parasitol,
41,
545-552.
|
 |
|
|
|
|
 |
S.Nagaoka,
Y.Takata,
and
K.Kato
(2011).
Identification of two arginases generated by alternative splicing in the silkworm, Bombyx mori.
|
| |
Arch Insect Biochem Physiol,
76,
97.
|
 |
|
|
|
|
 |
D.Schade,
J.Kotthaus,
and
B.Clement
(2010).
Modulating the NO generating system from a medicinal chemistry perspective: current trends and therapeutic options in cardiovascular disease.
|
| |
Pharmacol Ther,
126,
279-300.
|
 |
|
|
|
|
 |
L.Di Costanzo,
M.Ilies,
K.J.Thorn,
and
D.W.Christianson
(2010).
Inhibition of human arginase I by substrate and product analogues.
|
| |
Arch Biochem Biophys,
496,
101-108.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Sankaralingam,
H.Xu,
and
S.T.Davidge
(2010).
Arginase contributes to endothelial cell oxidative stress in response to plasma from women with preeclampsia.
|
| |
Cardiovasc Res,
85,
194-203.
|
 |
|
|
|
|
 |
H.Maarsingh,
J.Zaagsma,
and
H.Meurs
(2009).
Arginase: a key enzyme in the pathophysiology of allergic asthma opening novel therapeutic perspectives.
|
| |
Br J Pharmacol,
158,
652-664.
|
 |
|
|
|
|
 |
J.M.Fitzpatrick,
J.M.Fuentes,
I.W.Chalmers,
T.A.Wynn,
M.Modolell,
K.F.Hoffmann,
and
M.Hesse
(2009).
Schistosoma mansoni arginase shares functional similarities with human orthologs but depends upon disulphide bridges for enzymatic activity.
|
| |
Int J Parasitol,
39,
267-279.
|
 |
|
|
|
|
 |
N.Huynh,
E.Harris,
J.Chin-Dusting,
and
K.Andrews
(2009).
The vascular effects of different arginase inhibitors in rat isolated aorta and mesenteric arteries.
|
| |
Br J Pharmacol,
156,
84-93.
|
 |
|
|
|
|
 |
L.Santhanam,
D.W.Christianson,
D.Nyhan,
and
D.E.Berkowitz
(2008).
Arginase and vascular aging.
|
| |
J Appl Physiol,
105,
1632-1642.
|
 |
|
|
|
|
 |
T.Bagnost,
A.Berthelot,
M.Bouhaddi,
P.Laurant,
C.André,
Y.Guillaume,
and
C.Demougeot
(2008).
Treatment with the arginase inhibitor N(omega)-hydroxy-nor-L-arginine improves vascular function and lowers blood pressure in adult spontaneously hypertensive rat.
|
| |
J Hypertens,
26,
1110-1118.
|
 |
|
|
|
|
 |
L.Di Costanzo,
M.E.Pique,
and
D.W.Christianson
(2007).
Crystal structure of human arginase I complexed with thiosemicarbazide reveals an unusual thiocarbonyl mu-sulfide ligand in the binuclear manganese cluster.
|
| |
J Am Chem Soc,
129,
6388-6389.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.López,
R.Alarcón,
M.S.Orellana,
P.Enríquez,
E.Uribe,
J.Martínez,
and
N.Carvajal
(2005).
Insights into the interaction of human arginase II with substrate and manganese ions by site-directed mutagenesis and kinetic studies. Alteration of substrate specificity by replacement of Asn149 with Asp.
|
| |
FEBS J,
272,
4540-4548.
|
 |
|
|
|
|
 |
H.J.Ahn,
K.H.Kim,
J.Lee,
J.Y.Ha,
H.H.Lee,
D.Kim,
H.J.Yoon,
A.R.Kwon,
and
S.W.Suh
(2004).
Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily.
|
| |
J Biol Chem,
279,
50505-50513.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Ivanov,
and
M.L.Klein
(2004).
First principles computational study of the active site of arginase.
|
| |
Proteins,
54,
1-7.
|
 |
|
|
|
|
 |
L.Xu,
B.Hilliard,
R.J.Carmody,
G.Tsabary,
H.Shin,
D.W.Christianson,
and
Y.H.Chen
(2003).
Arginase and autoimmune inflammation in the central nervous system.
|
| |
Immunology,
110,
141-148.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

