 |
PDBsum entry 1hj8

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Atomic resolution structures of trypsin provide insight into structural radiation damage.
|
 |
|
Authors
|
 |
H.K.Leiros,
S.M.Mcsweeney,
A.O.Smalås.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2001,
57,
488-497.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
Radiation damage is an inherent problem in protein X-ray crystallography and the
process has recently been shown to be highly specific, exhibiting features such
as cleavage of disulfide bonds, decarboxylation of acidic residues, increase in
atomic B factors and increase in unit-cell volume. Reported here are two trypsin
structures at atomic resolution (1.00 and 0.95 A), the data for which were
collected at a third-generation synchrotron (ESRF) at two different beamlines.
Both trypsin structures exhibit broken disulfide bonds; in particular, the bond
from Cys191 to Cys220 is very sensitive to synchrotron radiation. The data set
collected at the most intense beamline (ID14-EH4) shows increased structural
radiation damage in terms of lower occupancies for cysteine residues, more
breakage in the six disulfide bonds and more alternate conformations. It appears
that high intensity and not only the total X-ray dose is most harmful to protein
crystals.
|
 |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 The catalytic hydrogen bond from His57 N 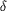 1
to Asp102 O 1
to Asp102 O 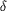 2
in anionic salmon trypsin (AST). The 2mF[o] - DF[c] map is
contoured at 2.0 2
in anionic salmon trypsin (AST). The 2mF[o] - DF[c] map is
contoured at 2.0  (blue)
and 4.0 (blue)
and 4.0  (red),
while the mF[o] - DF[c] map around the His57 side chain is
contoured at +2.5 (red),
while the mF[o] - DF[c] map around the His57 side chain is
contoured at +2.5  (green).
Ser195 and Ser214 are also included in the figure, which was
created with Bobscript (Esnouf, 1997[Esnouf, R. M. (1997). J.
Mol. Graph. 15, 132-134.]). (green).
Ser195 and Ser214 are also included in the figure, which was
created with Bobscript (Esnouf, 1997[Esnouf, R. M. (1997). J.
Mol. Graph. 15, 132-134.]).
|
 |
Figure 3.
Figure 3 (a) Stereoview of the salt-bridge interactions that
involve residues uniquely conserved in all cold-adapted trypsins
(Leiros et al., 1999[Leiros, H.-K. S., Willassen, N. P. &
Smalås, A. O. (1999). Extremophiles, 3, 205-219.], 2000[Leiros,
H.-K. S., Willassen, N. P. & Smalås, A. O. (2000). Eur. J.
Biochem. 267, 1039-1049.]). The 2mF[o] - DF[c] map of the side
chains only is contoured at 1.8  .
(b) Location of the ion-pair network relative to the N-terminal
(red), the calcium-binding loop (blue) and the autolysis loop
(yellow). .
(b) Location of the ion-pair network relative to the N-terminal
(red), the calcium-binding loop (blue) and the autolysis loop
(yellow).
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2001,
57,
488-497)
copyright 2001.
|
 |
|

|
|
|
 |

