 |
PDBsum entry 1fn8

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase substrate
|
PDB id
|
|

|

|
1fn8
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Fusarium oxysporum trypsin at atomic resolution at 100 and 283 k: a study of ligand binding.
|
 |
|
Authors
|
 |
W.R.Rypniewski,
P.R.Ostergaard,
M.Nørregaard-Madsen,
M.Dauter,
K.S.Wilson.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2001,
57,
8.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
The X-ray structure of F. oxysporum trypsin has been determined at atomic
resolution, revealing electron density in the binding site which was interpreted
as a peptide bound in the sites S1, S2 and S3. The structure, which was
initially determined at 1.07 A resolution and 283 K, has an Arg in the S1
specificity pocket. The study was extended to 0.81 A resolution at 100 K using
crystals soaked in Arg, Lys and Gln to study in greater detail the binding at
the S1 site. The electron density in the binding site was compared between the
different structures and analysed in terms of partially occupied and overlapping
components of peptide, solvent water and possibly other chemical moieties.
Arg-soaked crystals reveal a density more detailed but similar to the original
structure, with the Arg side chain visible in the S1 pocket and residual peptide
density in the S2 and S3 sites. The density in the active site is complex and
not fully interpreted. Lys at high concentrations displaces Arg in the S1
pocket, while some main-chain density remains in sites S2 and S3. Gln has been
shown not to bind. The free peptide in the S1-S3 sites binds in a similar way to
the binding loop of BPTI or the inhibitory domain of the Alzheimer's
beta-protein precursor, with some differences in the S1 site.
|
 |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Schematic representation of TRY-N. Secondary-structure
elements forming the N-terminal domain are marked A1-F1 and the
C-terminal domain A2-F2 as defined previously (Rypniewski,
Mangani et al., 1995[Rypniewski, W. R., Mangani, S., Bruni, B.,
Orioli, P. L., Casati, M. & Wilson, K. S. (1995). J. Mol. Biol.
251, 282-296.]). The active site is indicated by the side chains
of the `catalytic triad': Ser195, His57 and Asp102. The side
chain of Asp189 is also shown at the bottom of the `specificity
pocket' defining the tryptic substrate specificity. The peptide
bound in sites S1 to S3 is indicated by a ball-and-stick model
with dark bonds.
|
 |
Figure 6.
Figure 6 Ligand binding in TRY-LYS2. (a) `Omit' maps calculated
as for Fig. 4-(a), at the same contour levels and with similar
colouring scheme. Sites 2004-2006 are not occupied in this
structure, as shown by the absence of difference density. There
is an indication of a double conformation of the Lys side chain,
not modelled because of weak density level for refinement. The
water at 2102 has been modelled in two alternative
conformations: 2102b is at a bonding distance to Lys N 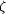 ,
whilst 2102a is at clashing distance, binding in the absence of
the peptide ligand. (b) Geometry of the binding site. The
labelling scheme is similar to Fig. 4-(b). Some atoms have been
omitted for clarity. ,
whilst 2102a is at clashing distance, binding in the absence of
the peptide ligand. (b) Geometry of the binding site. The
labelling scheme is similar to Fig. 4-(b). Some atoms have been
omitted for clarity.
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2001,
57,
8-0)
copyright 2001.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Structure of inhibited trypsin from fusarium oxysporum at 1.55 a.
|
 |
|
Authors
|
 |
W.R.Rypniewski,
C.Dambmann,
C.Von der osten,
M.Dauter,
K.S.Wilson.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 1995,
51,
73-85.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. Progress of the refinement: R factor
(r.[IFo I -
IFcl[/r~lFol)
against
the cycle number of the least-squares refinement. Dots indicate manual
rebuilding of the model. The point marked H indicates where the
contribution of H atoms was included in the refinement.
|
 |
Figure 11.
Fig. 11. The electron density
(3Fo
- 2Fc)
for the monoisopropyl
phosphoryl inhibitor group
bound to Ser195. Catalytic
His57
is shown on the right. Gln192,
which partly closes the binding
site is shown on the left. The
density for the side chain is
weak. The map was contoured
at the l tr level.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the IUCr
|
 |
|

|
|
|
 |

