 |
PDBsum entry 1e5d

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1e5d
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Structure of a dioxygen reduction enzyme from desulfovibrio gigas.
|
 |
|
Authors
|
 |
C.Frazão,
G.Silva,
C.M.Gomes,
P.Matias,
R.Coelho,
L.Sieker,
S.Macedo,
M.Y.Liu,
S.Oliveira,
M.Teixeira,
A.V.Xavier,
C.Rodrigues-Pousada,
M.A.Carrondo,
J.Le gall.
|
 |
|
Ref.
|
 |
Nat Struct Biol, 2000,
7,
1041-1045.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
Desulfovibrio gigas is a strict anaerobe that contains a well-characterized
metabolic pathway that enables it to survive transient contacts with oxygen. The
terminal enzyme in this pathway, rubredoxin:oxygen oxidoreductase (ROO) reduces
oxygen to water in a direct and safe way. The 2.5 A resolution crystal structure
of ROO shows that each monomer of this homodimeric enzyme consists of a novel
combination of two domains, a flavodoxin-like domain and a
Zn-beta-lactamase-like domain that contains a di-iron center for dioxygen
reduction. This is the first structure of a member of a superfamily of enzymes
widespread in strict and facultative anaerobes, indicating its broad
physiological significance.
|
 |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. ROO is a modular enzyme. a, The ROO dimer (monomers
in blue and brown), showing the  -lactamase-like
(light) and flavodoxin-like (dark) domains, iron (orange
spheres) and FMN (stick model). A two-fold axis relates both
monomers. b, The -lactamase-like
(light) and flavodoxin-like (dark) domains, iron (orange
spheres) and FMN (stick model). A two-fold axis relates both
monomers. b, The  -lactamase-like
domain. Left, a ribbon diagram with termini labeled. Right,
stereo view of the -lactamase-like
domain. Left, a ribbon diagram with termini labeled. Right,
stereo view of the  -lactamase-like
domain (blue) superimposed on -lactamase-like
domain (blue) superimposed on  -lactamases
from Stenotrophomonas maltophilia^11 (green), Bacillus cereus12
(deep pink) and Bacteroides fragilis13 (gold). Additional ROO
structural elements in the region corresponding to the substrate
groove of -lactamases
from Stenotrophomonas maltophilia^11 (green), Bacillus cereus12
(deep pink) and Bacteroides fragilis13 (gold). Additional ROO
structural elements in the region corresponding to the substrate
groove of  -lactamases
are indicated in dark blue. c, The flavodoxin-like domain. Left,
ribbon diagram. Right, stereo view of the flavodoxin-like domain
(blue) superimposed on Desulfovibrio vulgaris 32 (brown) and
Clostridium beijerinckii17 (violet) flavodoxins. -lactamases
are indicated in dark blue. c, The flavodoxin-like domain. Left,
ribbon diagram. Right, stereo view of the flavodoxin-like domain
(blue) superimposed on Desulfovibrio vulgaris 32 (brown) and
Clostridium beijerinckii17 (violet) flavodoxins.
|
 |
Figure 3.
Figure 3. 3 ROO catalytic site. a, Distances (Å) are given
for contacts to the pentacoordinated (quadrangular pyramid) Fe 1
and tetracoordinated (quadrangular plane) Fe 2 coordinations
(green), and for other close contacts (cyan). The oxygen
molecule (OXY) and water (WAT 1) are close to Fe 2, but outside
the typical bonding distances. Similar to other di-iron
proteins14, 15, this site contains a bridging  -O
(or -O
(or  -OH;
MUO) at hydrogen bonding distance from the Asp 83 OD1. The
electron density was contoured at 1 -OH;
MUO) at hydrogen bonding distance from the Asp 83 OD1. The
electron density was contoured at 1  (blue)
and 7 (blue)
and 7  (red).
b, ROO dimerization (monomers in blue and brown) couples the two
monomer cofactors. The spatial orientations of the side chains
of His 79, Glu 81, and His 226 are stabilized through hydrogen
bonds to the side chains of Asp 149 and Trp 263 and to the
carbonyl O of Tyr 34, respectively. Intradimer contacts involve
Trp 147, Pro 148 and Asp 149 as well as the aromatic ring of Trp
347. (red).
b, ROO dimerization (monomers in blue and brown) couples the two
monomer cofactors. The spatial orientations of the side chains
of His 79, Glu 81, and His 226 are stabilized through hydrogen
bonds to the side chains of Asp 149 and Trp 263 and to the
carbonyl O of Tyr 34, respectively. Intradimer contacts involve
Trp 147, Pro 148 and Asp 149 as well as the aromatic ring of Trp
347.
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2000,
7,
1041-1045)
copyright 2000.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Crystallization and preliminary diffraction data analysis of both single and pseudo-Merohedrally twinned crystals of rubredoxin oxygen oxidoreductase from desulfovibrio gigas.
|
 |
|
Authors
|
 |
C.Frazão,
L.Sieker,
R.Coelho,
J.Morais,
I.Pacheco,
L.Chen,
J.Legall,
Z.Dauter,
K.Wilson,
M.A.Carrondo.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 1999,
55,
1465-1467.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |
Figure 2.
Figure 2 Self-rotation function maps of ROO for (a) the 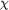 =
85° section and (b) the =
85° section and (b) the 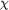 =
180° section. Non-crystallographic symmetry is recognized by
peak heights of 69% of the origin peak. The pseudo-fourfold
rotation axis (a) is parallel to c, with its maximum at =
180° section. Non-crystallographic symmetry is recognized by
peak heights of 69% of the origin peak. The pseudo-fourfold
rotation axis (a) is parallel to c, with its maximum at 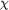 =
84.1°; the twofold rotation axis (b) lies at =
84.1°; the twofold rotation axis (b) lies at 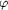 =
42.9, =
42.9,  =
90.0°. Map contouring is at 1 =
90.0°. Map contouring is at 1  . .
|
 |
|
 |
 |
|
The above figure is
reproduced from the cited reference
with permission from the IUCr
|
 |
|
Secondary reference #2
|
 |
|
Title
|
 |
Studies on the redox centers of the terminal oxidase from desulfovibrio gigas and evidence for its interaction with rubredoxin.
|
 |
|
Authors
|
 |
C.M.Gomes,
G.Silva,
S.Oliveira,
J.Legall,
M.Y.Liu,
A.V.Xavier,
C.Rodrigues-Pousada,
M.Teixeira.
|
 |
|
Ref.
|
 |
J Biol Chem, 1997,
272,
22502-22508.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
 |
 |

|
 |

|
 |
Figure 7.
Fig. 7. Interaction between reduced Rd and ROO. ROO (1.5
µM) was in 50 mM Tris-HCl buffer, pH 7.6. Inset, 16
µM oxidized Rd^ (Rd[ox]) was incubated anaerobically with
160 µM NADH and 3 nM NRO. After 5 min, full reduction was
achieved (Rd[red]). Trace a, native^ ROO; trace b, 1 min after
addition of reduced Rd (2 µM).
|
 |
Figure 8.
Fig. 8. Partial restriction map and predicted amino acid
sequences of Rd and ROO. Panel A, partial restriction map of the
subcloned^ 3.6-kilobase pair BamHI-BamHI DNA fragment. Panel B,
amino acid^ sequence of Rd and the N terminus of ROO.
|
 |
|
 |
 |
|
The above figures are
reproduced from the cited reference
with permission from the ASBMB
|
 |
|
Secondary reference #3
|
 |
|
Title
|
 |
Rubredoxin oxidase, A new flavo-Hemo-Protein, Is the site of oxygen reduction to water by the "strict anaerobe" desulfovibrio gigas.
|
 |
|
Authors
|
 |
L.Chen,
M.Y.Liu,
J.Legall,
P.Fareleira,
H.Santos,
A.V.Xavier.
|
 |
|
Ref.
|
 |
Biochem Biophys Res Commun, 1993,
193,
100-105.
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
|

|
|
|
 |

