 |
PDBsum entry 1uo6

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.21.36
- pancreatic elastase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Hydrolysis of proteins, including elastin. Preferential cleavage: Ala-|-Xaa.
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
60:28-38
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
On the routine use of soft X-rays in macromolecular crystallography. Part II. Data-collection wavelength and scaling models.
|
|
C.Mueller-Dieckmann,
M.Polentarutti,
K.Djinovic Carugo,
S.Panjikar,
P.A.Tucker,
M.S.Weiss.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Complete and highly redundant data sets were collected at nine different
wavelengths between 0.80 and 2.65 A on a xenon derivative of porcine pancreatic
elastase in both air and helium atmospheres. The magnitude of the anomalous
signal, as assessed by the xenon-peak height in the anomalous difference
Patterson synthesis, is affected by the wavelength of data collection as well as
by the scaling model used. For data collected at wavelengths longer than 1.7 A,
the use of a three-dimensional scaling protocol is essential in order to obtain
the highest possible anomalous signal. Based on the scaling protocols currently
available, the optimal wavelength range for data collection appears to be
between 2.1 and 2.4 A. Beyond that, any further increase in signal will be
compensated for or even superseded by a concomitant increase in noise, which
cannot be fully corrected for. Data collection in a helium atmosphere yields
higher I/sigma(I) values, but not significantly better anomalous differences,
than data collection in air.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Schematic representation of the relevant absorption
coefficients in a diffraction experiment. The four different
coefficients are 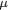 [1]
for absorption in the crystal, [1]
for absorption in the crystal, 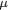 [2]
for absorption in the mother liquor surrounding the crystal, [2]
for absorption in the mother liquor surrounding the crystal,
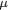 [3]
for absorption by the nylon loop used and [3]
for absorption by the nylon loop used and 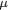 [4]
for absorption in air (or helium). [4]
for absorption in air (or helium).
|
 |
Figure 6.
Figure 6 I/  (I)
as a function of resolution (1/d^2) for the PPE-Xe data set
I-Air (black lines) and I-He (red lines). The dip at 3.5 Å
resolution in the upper curve is a result of the appearance of
an ice ring on the images of the later data sets. (I)
as a function of resolution (1/d^2) for the PPE-Xe data set
I-Air (black lines) and I-He (red lines). The dip at 3.5 Å
resolution in the upper curve is a result of the appearance of
an ice ring on the images of the later data sets.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2004,
60,
28-38)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Pompidor,
O.Maury,
J.Vicat,
and
R.Kahn
(2010).
A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
762-769.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

