 |
PDBsum entry 1qnj

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase (serine protease)
|
PDB id
|
|

|

|
1qnj
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.21.36
- pancreatic elastase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Hydrolysis of proteins, including elastin. Preferential cleavage: Ala-|-Xaa.
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
56:520-523
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Atomic resolution structure of native porcine pancreatic elastase at 1.1 A.
|
|
M.Würtele,
M.Hahn,
K.Hilpert,
W.Höhne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A data set from the serine protease porcine pancreatic elastase was collected at
atomic resolution (1.1 A) with synchrotron radiation. The improved resolution
allows the determination of atom positions with high accuracy, as well as the
localization of H atoms. Three residues could be modelled in alternative
positions. The catalytic triad of elastase consists of His57, Asp102 and Ser195.
The His57 N(delta1) H atom was located at a distance of 0.82 A from the
N(delta1) atom. The distance between His57 N(delta1) and Asp102 O(delta2) is
2.70 +/- 0.04 A, thus indicating normal hydrogen-bonding geometry. Additional H
atoms at His57 N(varepsilon2) and Ser195 O(gamma) could not be identified in the
F(o) - F(c) density maps.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 2.
Figure 2 The catalytic triad of PPE. (a) 2F[o] - F[c] map
contoured at 3.5  (blue)
shows atomic resolution of active-site residues of the final
model (SUL1: sulfate). (b) 2F[o] - F[c] map of anisotropic
B-factor model with the HN (blue)
shows atomic resolution of active-site residues of the final
model (SUL1: sulfate). (b) 2F[o] - F[c] map of anisotropic
B-factor model with the HN 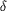 1
omitted contoured at 2.5 1
omitted contoured at 2.5  (blue).
F[o] - F[c] map is contoured at 2.5 (blue).
F[o] - F[c] map is contoured at 2.5  (red).
(c) Thermal ellipsoid representation of active-site residues was
made with ORTEX 7e (McArdle, 1995[McArdle, P. (1995). J. Appl.
Cryst. 28, 65.]; Burnett & Johnson, 1996[Burnett, M. N. &
Johnson, C. K. (1996). ORTEP III: Oak Ridge Thermal Ellipsoid
Plot Program for Crystal Structure Illustrations. Report
ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.]).
Putative hydrogen bonds are shown as dotted lines. (red).
(c) Thermal ellipsoid representation of active-site residues was
made with ORTEX 7e (McArdle, 1995[McArdle, P. (1995). J. Appl.
Cryst. 28, 65.]; Burnett & Johnson, 1996[Burnett, M. N. &
Johnson, C. K. (1996). ORTEP III: Oak Ridge Thermal Ellipsoid
Plot Program for Crystal Structure Illustrations. Report
ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.]).
Putative hydrogen bonds are shown as dotted lines.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2000,
56,
520-523)
copyright 2000.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Pompidor,
O.Maury,
J.Vicat,
and
R.Kahn
(2010).
A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
762-769.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.A.Sigala,
D.A.Kraut,
J.M.Caaveiro,
B.Pybus,
E.A.Ruben,
D.Ringe,
G.A.Petsko,
and
D.Herschlag
(2008).
Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
|
| |
J Am Chem Soc,
130,
13696-13708.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Vrielink,
and
N.Sampson
(2003).
Sub-Angstrom resolution enzyme X-ray structures: is seeing believing?
|
| |
Curr Opin Struct Biol,
13,
709-715.
|
 |
|
|
|
|
 |
G.Katona,
R.C.Wilmouth,
P.A.Wright,
G.I.Berglund,
J.Hajdu,
R.Neutze,
and
C.J.Schofield
(2002).
X-ray structure of a serine protease acyl-enzyme complex at 0.95-A resolution.
|
| |
J Biol Chem,
277,
21962-21970.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Overgaard,
B.Schiøtt,
F.K.Larsen,
and
B.B.Iversen
(2001).
The charge density distribution in a model compound of the catalytic triad in serine proteases.
|
| |
Chemistry,
7,
3756-3767.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

