 |
PDBsum entry 1odn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1odn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.21.3.1
- isopenicillin-N synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine + O2 = isopenicillin N + 2 H2O
|
 |
 |
 |
 |
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine
|
+
|
 O2
O2
|
=
|
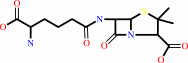 isopenicillin N
isopenicillin N
|
+
|
2
×
H2O
Bound ligand (Het Group name = )
matches with 92.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Org Biomol Chem
1:1455-1460
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic studies on the reaction of isopenicillin N synthase with an unsaturated substrate analogue.
|
|
J.M.Elkins,
P.J.Rutledge,
N.I.Burzlaff,
I.J.Clifton,
R.M.Adlington,
P.L.Roach,
J.E.Baldwin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Isopenicillin N synthase (IPNS) catalyses conversion of the linear tripeptide
delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-valine (ACV) to isopenicillin N
(IPN), the central step in biosynthesis of the beta-lactam antibiotics. The
unsaturated substrate analogue
delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-vinylglycine (ACvG) has previously
been incubated with IPNS and single product was isolated, a
2-alpha-hydroxymethyl isopenicillin N (HMPen), formed via a monooxygenase mode
of reactivity. ACvG has now been crystallised with IPNS and the structure of the
anaerobic IPNS:Fe(II):ACvG complex determined to 1.15 A resolution. Furthermore,
by exposing the anaerobically grown crystals to high-pressure oxygen gas, a
structure corresponding to the bicyclic product HMPen has been obtained at 1.60
A resolution. In light of these and other IPNS structures, and recent
developments with related dioxygenases, the [2 + 2] cycloaddition mechanism for
HMPen formation from ACvG has been revised, and a stepwise radical mechanism is
proposed. This revised mechanism remains consistent with the observed
stereospecificity of the transformation, but fits better with apparent
constraints on the coordination geometry around the active site iron atom.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
V.J.Dungan,
Y.Ortin,
H.Mueller-Bunz,
and
P.J.Rutledge
(2010).
Design and synthesis of a tetradentate '3-amine-1-carboxylate' ligand to mimic the metal binding environment at the non-heme iron(II) oxidase active site.
|
| |
Org Biomol Chem,
8,
1666-1673.
|
 |
|
|
|
|
 |
W.Ge,
I.J.Clifton,
A.R.Howard-Jones,
J.E.Stok,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2009).
Structural studies on the reaction of isopenicillin N synthase with a sterically demanding depsipeptide substrate analogue.
|
| |
Chembiochem,
10,
2025-2031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.C.Stewart,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2007).
A cyclobutanone analogue mimics penicillin in binding to isopenicillin N synthase.
|
| |
Chembiochem,
8,
2003-2007.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Daruzzaman,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2006).
Unexpected oxidation of a depsipeptide substrate analogue in crystalline isopenicillin N synthase.
|
| |
Chembiochem,
7,
351-358.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

