 |
PDBsum entry 1k9v

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.1.2
- glutaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutamine + H2O = L-glutamate + NH4+
|
 |
 |
 |
 |
 |
 L-glutamine
L-glutamine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
 L-glutamate
L-glutamate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.4.3.2.10
- imidazole glycerol-phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide + L-glutamine = D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate + 5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide + L-glutamate + H+
|
 |
 |
 |
 |
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide
|
+
|
L-glutamine
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
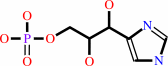 D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
|
+
|
5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamate
L-glutamate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
10:185-193
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
|
|
A.Douangamath,
M.Walker,
S.Beismann-Driemeyer,
M.C.Vega-Fernandez,
R.Sterner,
M.Wilmanns.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Since reactive ammonia is not available under physiological conditions,
glutamine is used as a source for the incorporation of nitrogen in a number of
metabolic pathway intermediates. The heterodimeric ImGP synthase that links
histidine and purine biosynthesis belongs to the family of glutamine
amidotransferases in which the glutaminase activity is coupled with a subsequent
synthase activity specific for each member of the enzyme family. Its X-ray
structure from the hyperthermophile Thermotoga maritima shows that the
glutaminase subunit is associated with the N-terminal face of the (beta
alpha)(8) barrel cyclase subunit. The complex reveals a putative tunnel for the
transfer of ammonia over a distance of 25 A. Although ammonia tunneling has been
reported for glutamine amidotransferases, the ImGP synthase has evolved a novel
mechanism, which extends the known functional properties of the versatile (beta
alpha)(8) barrel fold.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. Reactions Catalyzed by the Glutaminase Subunit
HisH and the Cyclase Subunit HisF, which Constitute the ImGP
SynthaseThe products ImGP and AICAR are further used in
histidine and de novo purine biosynthesis, respectively.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2002,
10,
185-193)
copyright 2002.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Liebold,
F.List,
H.R.Kalbitzer,
R.Sterner,
and
E.Brunner
(2010).
The interaction of ammonia and xenon with the imidazole glycerol phosphate synthase from Thermotoga maritima as detected by NMR spectroscopy.
|
| |
Protein Sci,
19,
1774-1782.
|
 |
|
|
|
|
 |
J.M.Lipchock,
and
J.P.Loria
(2010).
Nanometer propagation of millisecond motions in V-type allostery.
|
| |
Structure,
18,
1596-1607.
|
 |
|
|
|
|
 |
L.Lund,
Y.Fan,
Q.Shao,
Y.Q.Gao,
and
F.M.Raushel
(2010).
Carbamate transport in carbamoyl phosphate synthetase: a theoretical and experimental investigation.
|
| |
J Am Chem Soc,
132,
3870-3878.
|
 |
|
|
|
|
 |
J.Lipchock,
and
J.P.Loria
(2009).
Millisecond dynamics in the allosteric enzyme imidazole glycerol phosphate synthase (IGPS) from Thermotoga maritima.
|
| |
J Biomol NMR,
45,
73-84.
|
 |
|
|
|
|
 |
R.Koike,
A.Kidera,
and
M.Ota
(2009).
Alteration of oligomeric state and domain architecture is essential for functional transformation between transferase and hydrolase with the same scaffold.
|
| |
Protein Sci,
18,
2060-2066.
|
 |
|
|
|
|
 |
Y.Fan,
L.Lund,
Q.Shao,
Y.Q.Gao,
and
F.M.Raushel
(2009).
A combined theoretical and experimental study of the ammonia tunnel in carbamoyl phosphate synthetase.
|
| |
J Am Chem Soc,
131,
10211-10219.
|
 |
|
|
|
|
 |
E.J.Hart,
and
S.G.Powers-Lee
(2008).
Mutation analysis of carbamoyl phosphate synthetase: does the structurally conserved glutamine amidotransferase triad act as a functional dyad?
|
| |
Protein Sci,
17,
1120-1128.
|
 |
|
|
|
|
 |
J.M.Lipchock,
and
J.P.Loria
(2008).
1H, 15N and 13C resonance assignment of imidazole glycerol phosphate (IGP) synthase protein HisF from Thermotoga maritima.
|
| |
Biomol NMR Assign,
2,
219-221.
|
 |
|
|
|
|
 |
M.A.Vanoni,
and
B.Curti
(2008).
Structure-function studies of glutamate synthases: a class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation.
|
| |
IUBMB Life,
60,
287-300.
|
 |
|
|
|
|
 |
M.T.Reetz,
M.Rentzsch,
A.Pletsch,
A.Taglieber,
F.Hollmann,
R.J.Mondière,
N.Dickmann,
B.Höcker,
S.Cerrone,
M.C.Haeger,
and
R.Sterner
(2008).
A robust protein host for anchoring chelating ligands and organocatalysts.
|
| |
Chembiochem,
9,
552-564.
|
 |
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Conformational changes in ammonia-channeling glutamine amidotransferases.
|
| |
Curr Opin Struct Biol,
17,
653-664.
|
 |
|
|
|
|
 |
A.Nakamura,
M.Yao,
S.Chimnaronk,
N.Sakai,
and
I.Tanaka
(2006).
Ammonia channel couples glutaminase with transamidase reactions in GatCAB.
|
| |
Science,
312,
1954-1958.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Zein,
Y.Zhang,
Y.N.Kang,
K.Burns,
T.P.Begley,
and
S.E.Ealick
(2006).
Structural insights into the mechanism of the PLP synthase holoenzyme from Thermotoga maritima.
|
| |
Biochemistry,
45,
14609-14620.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Quevillon-Cheruel,
N.Leulliot,
M.Graille,
K.Blondeau,
J.Janin,
and
H.van Tilbeurgh
(2006).
Crystal structure of the yeast His6 enzyme suggests a reaction mechanism.
|
| |
Protein Sci,
15,
1516-1521.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.A.Vanoni,
L.Dossena,
R.H.van den Heuvel,
and
B.Curti
(2005).
Structure-function studies on the complex iron-sulfur flavoprotein glutamate synthase: the key enzyme of ammonia assimilation.
|
| |
Photosynth Res,
83,
219-238.
|
 |
|
|
|
|
 |
R.E.Amaro,
R.S.Myers,
V.J.Davisson,
and
Z.A.Luthey-Schulten
(2005).
Structural elements in IGP synthase exclude water to optimize ammonia transfer.
|
| |
Biophys J,
89,
475-487.
|
 |
|
|
|
|
 |
J.A.Bauer,
E.M.Bennett,
T.P.Begley,
and
S.E.Ealick
(2004).
Three-dimensional structure of YaaE from Bacillus subtilis, a glutaminase implicated in pyridoxal-5'-phosphate biosynthesis.
|
| |
J Biol Chem,
279,
2704-2711.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Goto,
R.Omi,
N.Nakagawa,
I.Miyahara,
and
K.Hirotsu
(2004).
Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites.
|
| |
Structure,
12,
1413-1423.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Matte,
J.Sivaraman,
I.Ekiel,
K.Gehring,
Z.Jia,
and
M.Cygler
(2003).
Contribution of structural genomics to understanding the biology of Escherichia coli.
|
| |
J Bacteriol,
185,
3994-4002.
|
 |
|
|
|
|
 |
O.Mayans,
A.Ivens,
L.J.Nissen,
K.Kirschner,
and
M.Wilmanns
(2002).
Structural analysis of two enzymes catalysing reverse metabolic reactions implies common ancestry.
|
| |
EMBO J,
21,
3245-3254.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

