 |
PDBsum entry 1hb1

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Antibiotic biosynthesis
|
PDB id
|
|

|

|
1hb1
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.21.3.1
- isopenicillin-N synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine + O2 = isopenicillin N + 2 H2O
|
 |
 |
 |
 |
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine
|
+
|
 O2
O2
|
=
|
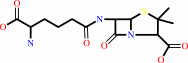 isopenicillin N
isopenicillin N
|
+
|
2
×
H2O
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Chem Biol
8:1231-1237
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Alternative oxidation by isopenicillin N synthase observed by X-ray diffraction.
|
|
J.M.Ogle,
I.J.Clifton,
P.J.Rutledge,
J.M.Elkins,
N.I.Burzlaff,
R.M.Adlington,
P.L.Roach,
J.E.Baldwin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Isopenicillin N synthase (IPNS) catalyses formation of bicyclic
isopenicillin N, precursor to all penicillin and cephalosporin antibiotics, from
the linear tripeptide delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-valine. IPNS is
a non-haem iron(II)-dependent enzyme which utilises the full oxidising potential
of molecular oxygen in catalysing the bicyclisation reaction. The reaction
mechanism is believed to involve initial formation of the beta-lactam ring (via
a thioaldehyde intermediate) to give an iron(IV)-oxo species, which then
mediates closure of the 5-membered thiazolidine ring. RESULTS: Here we report
experiments employing time-resolved crystallography to observe turnover of an
isosteric substrate analogue designed to intercept the catalytic pathway at an
early stage. Reaction in the crystalline enzyme-substrate complex was initiated
by the application of high-pressure oxygen, and subsequent flash freezing
allowed an oxygenated product to be trapped, bound at the iron centre. A
mechanism for formation of the observed thiocarboxylate product is proposed.
CONCLUSIONS: In the absence of its natural reaction partner (the N-H proton of
the L-cysteinyl-D-valine amide bond), the proposed hydroperoxide intermediate
appears to attack the putative thioaldehyde species directly. These results shed
light on the events preceding beta-lactam closure in the IPNS reaction cycle,
and enhance our understanding of the mechanism for reaction of the enzyme with
its natural substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Mechanisms for the reaction of IPNS with ACV and
ACOV. (a) Proposed mechanism for the reaction of IPNS with its
natural substrate ACV [3]. (b) Proposed pathway for generation
of the thiocarboxylate product from the substrate analogue ACOV.
See text for details of compounds 1–10. AA, δ-( Image
-α-aminoadipoyl).
|
 |
Figure 4.
Fig. 4. Potential hydrogen bonding interactions in the
active site region of the exposed IPNS:Fe(II):ACOV structure.
The three exposed structures oriented to show the potential for
hydrogen bonding around the thiocarboxylate oxygen, and to
demonstrate the consistency of the thiocarboxylate electron
density across the three structures. (a) From the 1.30 Å
resolution structure in the plane of the thiocarboxylate,
showing distances to Wat 402 and Wat 403. (b) The 1.30 Å
30 s, (c) the 1.40 Å 30 s, and (d) the 1.50 Å 120 s
structures from an alternative angle.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Chem Biol
(2001,
8,
1231-1237)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.D.Brown-Marshall,
A.R.Diebold,
and
E.I.Solomon
(2010).
Reaction coordinate of isopenicillin N synthase: oxidase versus oxygenase activity.
|
| |
Biochemistry,
49,
1176-1182.
|
 |
|
|
|
|
 |
V.J.Dungan,
Y.Ortin,
H.Mueller-Bunz,
and
P.J.Rutledge
(2010).
Design and synthesis of a tetradentate '3-amine-1-carboxylate' ligand to mimic the metal binding environment at the non-heme iron(II) oxidase active site.
|
| |
Org Biomol Chem,
8,
1666-1673.
|
 |
|
|
|
|
 |
W.Ge,
I.J.Clifton,
A.R.Howard-Jones,
J.E.Stok,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2009).
Structural studies on the reaction of isopenicillin N synthase with a sterically demanding depsipeptide substrate analogue.
|
| |
Chembiochem,
10,
2025-2031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.C.Stewart,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2007).
A cyclobutanone analogue mimics penicillin in binding to isopenicillin N synthase.
|
| |
Chembiochem,
8,
2003-2007.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.M.Bollinger,
and
C.Krebs
(2007).
Enzymatic C-H activation by metal-superoxo intermediates.
|
| |
Curr Opin Chem Biol,
11,
151-158.
|
 |
|
|
|
|
 |
A.Daruzzaman,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2006).
Unexpected oxidation of a depsipeptide substrate analogue in crystalline isopenicillin N synthase.
|
| |
Chembiochem,
7,
351-358.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

