 |
PDBsum entry 6ucn
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.3.1
- steroid Delta-isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 3-oxo-Delta5-steroid = a 3-oxo-Delta4-steroid
|
 |
 |
 |
 |
 |
3-oxo-Delta(5)-steroid
Bound ligand (Het Group name = )
matches with 90.48% similarity
|
=
|
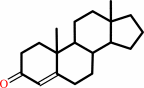 3-oxo-Delta(4)-steroid
3-oxo-Delta(4)-steroid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
117:33204-33215
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
|
|
F.Yabukarski,
J.T.Biel,
M.M.Pinney,
T.Doukov,
A.S.Powers,
J.S.Fraser,
D.Herschlag.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
How enzymes achieve their enormous rate enhancements remains a central question
in biology, and our understanding to date has impacted drug development,
influenced enzyme design, and deepened our appreciation of evolutionary
processes. While enzymes position catalytic and reactant groups in active sites,
physics requires that atoms undergo constant motion. Numerous proposals have
invoked positioning or motions as central for enzyme function, but a scarcity of
experimental data has limited our understanding of positioning and motion, their
relative importance, and their changes through the enzyme's reaction cycle. To
examine positioning and motions and test catalytic proposals, we collected
"room temperature" X-ray crystallography data for Pseudomonas
putida ketosteroid isomerase (KSI), and we obtained conformational ensembles
for this and a homologous KSI from multiple PDB crystal structures. Ensemble
analyses indicated limited change through KSI's reaction cycle. Active site
positioning was on the 1- to 1.5-Å scale, and was not exceptional compared to
noncatalytic groups. The KSI ensembles provided evidence against catalytic
proposals invoking oxyanion hole geometric discrimination between the ground
state and transition state or highly precise general base positioning. Instead,
increasing or decreasing positioning of KSI's general base reduced catalysis,
suggesting optimized Ångstrom-scale conformational heterogeneity that allows
KSI to efficiently catalyze multiple reaction steps. Ensemble analyses of
surrounding groups for WT and mutant KSIs provided insights into the forces and
interactions that allow and limit active-site motions. Most generally, this
ensemble perspective extends traditional structure-function relationships,
providing the basis for a new era of "ensemble-function" interrogation
of enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

