 |
PDBsum entry 6r3q

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Membrane protein
|
PDB id
|
|

|

|
6r3q
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chain A:
E.C.4.6.1.1
- adenylate cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ATP = 3',5'-cyclic AMP + diphosphate
|
 |
 |
 |
 |
 |
ATP
Bound ligand (Het Group name = )
matches with 90.91% similarity
|
=
|
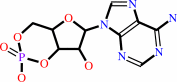 3',5'-cyclic AMP
3',5'-cyclic AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 2:
|
 |
Chain B:
E.C.?
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
364:389-394
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein.
|
|
C.Qi,
S.Sorrentino,
O.Medalia,
V.M.Korkhov.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Membrane-integral adenylyl cyclases (ACs) are key enzymes in mammalian
heterotrimeric GTP-binding protein (G protein)-dependent signal transduction,
which is important in many cellular processes. Signals received by the G
protein-coupled receptors are conveyed to ACs through G proteins to modulate the
levels of cellular cyclic adenosine monophosphate (cAMP). Here, we describe the
cryo-electron microscopy structure of the bovine membrane AC9 bound to an
activated G protein αs subunit at 3.4-angstrom resolution. The structure
reveals the organization of the membrane domain and helical domain that spans
between the membrane and catalytic domains of AC9. The carboxyl-terminal
extension of the catalytic domain occludes both the catalytic and the allosteric
sites of AC9, inducing a conformation distinct from the substrate- and
activator-bound state, suggesting a regulatory role in cAMP production.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

