 |
PDBsum entry 6qnh
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.5
- xylose isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-xylose = alpha-D-xylulofuranose
|
 |
 |
 |
 |
 |
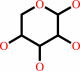 alpha-D-xylose
alpha-D-xylose
|
=
|
alpha-D-xylulofuranose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Methods
16:979-982
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Liquid application method for time-resolved analyses by serial synchrotron crystallography.
|
|
P.Mehrabi,
E.C.Schulz,
M.Agthe,
S.Horrell,
G.Bourenkov,
D.von Stetten,
J.P.Leimkohl,
H.Schikora,
T.R.Schneider,
A.R.Pearson,
F.Tellkamp,
R.J.D.Miller.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We introduce a liquid application method for time-resolved analyses (LAMA), an
in situ mixing approach for serial crystallography. Picoliter-sized droplets are
shot onto chip-mounted protein crystals, achieving near-full ligand occupancy
within theoretical diffusion times. We demonstrate proof-of-principle binding of
GlcNac to lysozyme, and resolve glucose binding and subsequent ring opening in a
time-resolved study of xylose isomerase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

