 |
PDBsum entry 6i6f

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
6i6f
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.153
- sepiapterin reductase (L-erythro-7,8-dihydrobiopterin forming).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biopterin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-erythro-7,8-dihydrobiopterin + NADP+ = L-sepiapterin + NADPH + H+
|
|
2.
|
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin + 2 NADP+ = 6-pyruvoyl- 5,6,7,8-tetrahydropterin + 2 NADPH + 2 H+
|
|
 |
 |
 |
 |
 |
L-erythro-7,8-dihydrobiopterin
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
L-sepiapterin
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin
|
+
|
2
×
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
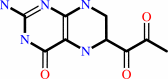 6-pyruvoyl- 5,6,7,8-tetrahydropterin
6-pyruvoyl- 5,6,7,8-tetrahydropterin
|
+
|
 2
×
NADPH
2
×
NADPH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
62:6391-6397
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
|
|
J.Alen,
M.Schade,
M.Wagener,
F.Christian,
S.Nordhoff,
B.Merla,
T.R.Dunkern,
G.Bahrenberg,
P.Ratcliffe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Genome-wide-association studies in chronic low back pain patients identified
sepiapterin reductase as a high interest target for developing new analgesics.
Here we used 19F NMR fragment screening for the discovery of novel,
ligand-efficient SPR inhibitors. We report the crystal structures of six
chemically diverse inhibitors complexed with SPR, identifying relevant
interactions and binding modes in the sepiapterin pocket. Exploration of our
initial fragment screening hit led to double-digit nanomolar inhibitors of SPR
with excellent ligand efficiency.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

