 |
PDBsum entry 6fvh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Macrophage migration inhibitory factor (mif) with covalently bound pitc
|
|
Structure:
|
 |
Macrophage migration inhibitory factor. Chain: a, b, c. Synonym: mif,glycosylation-inhibiting factor,gif,l-dopachrome isomerase,l-dopachrome tautomerase,phenylpyruvate tautomerase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: mif, glif, mmif. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.40Å
|
R-factor:
|
0.139
|
R-free:
|
0.166
|
|
|
Authors:
|
 |
V.R.Samygina,G.Bourenkov,A.V.Sokolov
|
|
Key ref:
|
 |
A.V.Sokolov
et al.
(2018).
Structural Study of the Complex Formed by Ceruloplasmin and Macrophage Migration Inhibitory Factor.
Biochemistry (Mosc),
83,
701-707.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-Mar-18
|
Release date:
|
06-Jun-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14174
(MIF_HUMAN) -
Macrophage migration inhibitory factor from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
115 a.a.
114 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.3.2.1
- phenylpyruvate tautomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-phenylpyruvate = enol-phenylpyruvate
|
 |
 |
 |
 |
 |
3-phenylpyruvate
|
=
|
enol-phenylpyruvate
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.3.12
- L-dopachrome isomerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-dopachrome = 5,6-dihydroxyindole-2-carboxylate
|
 |
 |
 |
 |
 |
L-dopachrome
Bound ligand (Het Group name = )
matches with 53.33% similarity
|
=
|
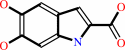 5,6-dihydroxyindole-2-carboxylate
5,6-dihydroxyindole-2-carboxylate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry (Mosc)
83:701-707
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural Study of the Complex Formed by Ceruloplasmin and Macrophage Migration Inhibitory Factor.
|
|
A.V.Sokolov,
L.A.Dadinova,
M.V.Petoukhov,
G.Bourenkov,
K.M.Dubova,
S.V.Amarantov,
V.V.Volkov,
V.A.Kostevich,
N.P.Gorbunov,
N.A.Grudinina,
V.B.Vasilyev,
V.R.Samygina.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Macrophage migration inhibitory factor (MIF) is a key proinflammatory cytokine.
Inhibitors of tautomerase activity of MIF are perspective antiinflammatory
compounds. Ceruloplasmin, the copper-containing ferroxidase of blood plasma, is
a noncompetitive inhibitor of tautomerase activity of MIF in the reaction with
p-hydroxyphenylpyruvate. Small-angle X-ray scattering established a model of the
complex formed by MIF and ceruloplasmin. Crystallographic analysis of MIF with a
modified active site supports the model. The stoichiometry of 3 CP/MIF trimer
complex was established using gel filtration. Conformity of novel data
concerning the interaction regions in the studied proteins with previous
biochemical data is discussed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

