 |
PDBsum entry 5ypp
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of ilvn.Val-1a
|
|
Structure:
|
 |
Acetolactate synthase isozyme 1 small subunit. Chain: a, b, c, d, e, f. Synonym: acetohydroxy-acid synthase i small subunit,als-i. Engineered: yes
|
|
Source:
|
 |
Escherichia coli (strain k12). Organism_taxid: 83333. Strain: k12. Gene: ilvn, b3670, jw3645. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.150
|
R-free:
|
0.170
|
|
|
Authors:
|
 |
S.P.Sarma,A.Bansal,H.Schindelin,B.Demeler
|
|
Key ref:
|
 |
A.Bansal
et al.
(2019).
Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry,
58,
1992-2008.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-Nov-17
|
Release date:
|
19-Sep-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0ADF8
(ILVN_ECOLI) -
Acetolactate synthase isozyme 1 small subunit from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
96 a.a.
90 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.6
- acetolactate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Isoleucine and Valine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 pyruvate + H+ = (2S)-2-acetolactate + CO2
|
 |
 |
 |
 |
 |
 2
×
pyruvate
2
×
pyruvate
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 55.56% similarity
|
=
|
(2S)-2-acetolactate
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 75.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
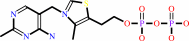 Thiamine diphosphate
Thiamine diphosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
58:1992-2008
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
|
|
A.Bansal,
N.M.Karanth,
B.Demeler,
H.Schindelin,
S.P.Sarma.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Conformational factors that predicate selectivity for valine or isoleucine
binding to IlvN leading to the regulation of aceto hydroxy acid synthase I (AHAS
I) of Escherichia coli have been determined for the first time from
high-resolution (1.9-2.43 Å) crystal structures of IlvN·Val and IlvN·Ile
complexes. The valine and isoleucine ligand binding pockets are located at the
dimer interface. In the IlvN·Ile complex, among residues in the binding pocket,
the side chain of Cys43 is 2-fold disordered (χ1 angles
of gauche- and trans). Only one conformation can be observed for the
identical residue in the IlvN·Val complexes. In a reversal, the side chain of
His53, located at the surface of the protein, exhibits two
conformations in the IlvN·Val complex. The concerted conformational switch in
the side chains of Cys43 and His53 may play an important
role in the regulation of the AHAS I holoenzyme activity. A significant result
is the establishment of the subunit composition in the AHAS I holoenzyme by
analytical ultracentrifugation. Solution nuclear magnetic resonance and
analytical ultracentrifugation experiments have also provided important insights
into the hydrodynamic properties of IlvN in the ligand-free and -bound states.
The structural and biophysical data unequivocally establish the molecular basis
for differential binding of the ligands to IlvN and a rationale for the
resistance of IlvM to feedback inhibition by the branched-chain amino acids.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

