 |
PDBsum entry 5tmp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.4.9
- dTMP kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dTMP + ATP = dTDP + ADP
|
 |
 |
 |
 |
 |
dTMP
Bound ligand (Het Group name = )
matches with 54.39% similarity
|
+
|
 ATP
ATP
|
=
|
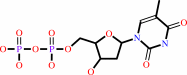 dTDP
dTDP
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
95:14045-14050
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for efficient phosphorylation of 3'-azidothymidine monophosphate by Escherichia coli thymidylate kinase.
|
|
A.Lavie,
N.Ostermann,
R.Brundiers,
R.S.Goody,
J.Reinstein,
M.Konrad,
I.Schlichting.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of Escherichia coli thymidylate kinase (TmpK) in complex
with P1-(5'-adenosyl)-P5-(5'-thymidyl)pentaphosphate and
pentaphosphate have been
solved to 2.0-A and 2.2-A resolution, respectively. The overall structure of the
bacterial TmpK is very similar to that of yeast TmpK. In contrast to the human
and yeast TmpKs, which phosphorylate 3'-azido-3'-deoxythymidine 5'-monophosphate
(AZT-MP) at a 200-fold reduced turnover number (kcat) in comparison to the
physiological substrate dTMP, reduction of kcat is only 2-fold for the bacterial
enzyme. The different kinetic properties toward AZT-MP between the eukaryotic
TmpKs and E. coli TmpK can be rationalized by the different ways in which these
enzymes stabilize the presumed transition state and the different manner in
which a carboxylic acid side chain in the P loop interacts with the deoxyribose
of the monophosphate. Yeast TmpK interacts with the 3'-hydroxyl of dTMP through
Asp-14 of the P loop in a bidentate manner: binding of AZT-MP results in a shift
of the P loop to accommodate the larger substituent. In E. coli TmpK, the
corresponding residue is Glu-12, and it interacts in a side-on fashion with the
3'-hydroxyl of dTMP. This different mode of interaction between the P loop
carboxylic acid with the 3' substituent of the monophosphate deoxyribose allows
the accommodation of an azido group in the case of the E. coli enzyme without
significant P loop movement. In addition, although the yeast enzyme uses Arg-15
(a glycine in E. coli) to stabilize the transition state, E. coli seems to use
Arg-153 from a region termed Lid instead. Thus, the binding of AZT-MP to the
yeast TmpK results in the shift of a catalytic residue, which is not the case
for the bacterial kinase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Interactions of the bisubstrate inhibitor with
TmpK (a). Distance map of TP[5]A bound to TmpK[coli]. P loop
residues are marked with an asterisk. (b-d) Stereoviews. Overlay
of the TmpK[coli]-TP[5]A complex model (pink) with the
TmpK[yeast]-TP[5]A model (green) (b and c) or the
TmpK[coli]-AZTP[5]A (blue) (d). (b) Interactions of the
3'-hydroxyl of the thymidine deoxyribose. In TmpK[yeast], a
bidentate interaction between the P loop aspartic acid and the
sugar hydroxyl is observed. The binding of AZT-MP causes the P
loop to move, thus displacing the catalytic P loop arginine. In
contrast, in TmpK[coli], the interaction between Glu-12 and the
3'-hydroxyl is side-on, and the bulkier azido group does not
induce a significant movement of the P loop. (c) Similar
phosphate-arginine interactions made in TmpK[yeast] by Arg-15
and in TmpK[coli] by Arg-153. Displayed are the P loop and a
part of the Lid region. The structures were overlaid according
to the position of the bisubstrate inhibitor. (d) In the
TmpK[coli]-TP[5]A and the TmpK[coli]-AZTP[5]A complex
structures, the thymine base is at an identical position, but
the deoxyribose moiety has undergone a rigid-body rotation
caused by the azido group in the AZT-P[5]A complex. In addition,
Glu-12 has rotated slightly to provide more room for the azido
group. The rotation of the deoxyribose induces a similar
rotation of the Glu-160 side chain. As Glu-12 makes close
interactions with Asp-157, the latter carboxylic acid also
rotates slightly. b-d were generated by using BOBSCRIPT (29, 30)
and RASTER 3D (31).
|
 |
Figure 2.
Fig. 2. Structure-based sequence alignment of the P loop
and Lid regions. Lysine and arginine residues that make
phosphate interactions are underlined doubly and those that make
a stacking interaction with the adenine base are underlined
singly. TmpKs are unique in having a carboxylic acid situated at
the tip of the P loop (Glu-12 in TmpK[coli]). In type I TmpKs
(e.g., human and yeast), the following residue is an arginine
that has been shown to be catalytically important for the yeast
enzyme. Type II TmpKs (e.g., E. coli) lack this arginine, having
instead a number of basic residues in their Lid region: for
TmpK[coli], Arg-153 presumably fulfills a catalytically role
analogous to that of Arg-15 in yeast. Although the last Lid
arginine in TmpKs (Arg-158 in TmpK[coli]) aligns well with
catalytic arginines from pig adenylate kinase (AK[pig]) and
Dictyostelium uridylate kinase (UmpK[dicty]), it points away
from the active site, thus having no obvious catalytic role. The
eukaryotic TmpKs have no catalytic residues in the Lid region.
|
 |
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.L.Whittingham,
J.Carrero-Lerida,
J.A.Brannigan,
L.M.Ruiz-Perez,
A.P.Silva,
M.J.Fogg,
A.J.Wilkinson,
I.H.Gilbert,
K.S.Wilson,
and
D.González-Pacanowska
(2010).
Structural basis for the efficient phosphorylation of AZT-MP (3'-azido-3'-deoxythymidine monophosphate) and dGMP by Plasmodium falciparum type I thymidylate kinase.
|
| |
Biochem J,
428,
499-509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Gogolin,
R.Seidel,
M.Engelhard,
R.S.Goody,
and
C.F.Becker
(2010).
Semisynthesis of human thymidine monophosphate kinase.
|
| |
Biopolymers,
94,
433-440.
|
 |
|
|
|
|
 |
M.Kandeel,
T.Ando,
Y.Kitamura,
M.Abdel-Aziz,
and
Y.Kitade
(2009).
Mutational, inhibitory and microcalorimetric analyses of Plasmodium falciparum TMP kinase. Implications for drug discovery.
|
| |
Parasitology,
136,
11-25.
|
 |
|
|
|
|
 |
S.Lutz,
L.Liu,
and
Y.Liu
(2009).
Engineering Kinases to Phosphorylate Nucleoside Analogs for Antiviral and Cancer Therapy.
|
| |
Chimia (Aarau),
63,
737-744.
|
 |
|
|
|
|
 |
A.Lavie,
Y.Su,
M.Ghassemi,
R.M.Novak,
M.Caffrey,
N.Sekulic,
C.Monnerjahn,
M.Konrad,
and
J.L.Cook
(2008).
Restoration of the antiviral activity of 3'-azido-3'-deoxythymidine (AZT) against AZT-resistant human immunodeficiency virus by delivery of engineered thymidylate kinase to T cells.
|
| |
J Gen Virol,
89,
1672-1679.
|
 |
|
|
|
|
 |
A.Ronceret,
J.Gadea-Vacas,
J.Guilleminot,
F.Lincker,
V.Delorme,
S.Lahmy,
G.Pelletier,
M.E.Chabouté,
and
M.Devic
(2008).
The first zygotic division in Arabidopsis requires de novo transcription of thymidylate kinase.
|
| |
Plant J,
53,
776-789.
|
 |
|
|
|
|
 |
B.Nocek,
S.Kochinyan,
M.Proudfoot,
G.Brown,
E.Evdokimova,
J.Osipiuk,
A.M.Edwards,
A.Savchenko,
A.Joachimiak,
and
A.F.Yakunin
(2008).
Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
|
| |
Proc Natl Acad Sci U S A,
105,
17730-17735.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Carnrot,
L.Wang,
D.Topalis,
and
S.Eriksson
(2008).
Mechanisms of substrate selectivity for Bacillus anthracis thymidylate kinase.
|
| |
Protein Sci,
17,
1486-1493.
|
 |
|
|
|
|
 |
M.Kandeel,
and
Y.Kitade
(2008).
Molecular characterization, heterologous expression and kinetic analysis of recombinant Plasmodium falciparum thymidylate kinase.
|
| |
J Biochem,
144,
245-250.
|
 |
|
|
|
|
 |
A.Ofiteru,
N.Bucurenci,
E.Alexov,
T.Bertrand,
P.Briozzo,
H.Munier-Lehmann,
and
A.M.Gilles
(2007).
Structural and functional consequences of single amino acid substitutions in the pyrimidine base binding pocket of Escherichia coli CMP kinase.
|
| |
FEBS J,
274,
3363-3373.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Khan,
S.Xiang,
and
L.Tong
(2007).
Crystal structure of human nicotinamide riboside kinase.
|
| |
Structure,
15,
1005-1013.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Hible,
P.Christova,
L.Renault,
E.Seclaman,
A.Thompson,
E.Girard,
H.Munier-Lehmann,
and
J.Cherfils
(2006).
Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase.
|
| |
Proteins,
62,
489-500.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.von Müller,
C.Schliep,
M.Storck,
W.Hampl,
T.Schmid,
D.Abendroth,
and
T.Mertens
(2006).
Severe graft rejection, increased immunosuppression, and active CMV infection in renal transplantation.
|
| |
J Med Virol,
78,
394-399.
|
 |
|
|
|
|
 |
M.Kotaka,
B.Dhaliwal,
J.Ren,
C.E.Nichols,
R.Angell,
M.Lockyer,
A.R.Hawkins,
and
D.K.Stammers
(2006).
Structures of S. aureus thymidylate kinase reveal an atypical active site configuration and an intermediate conformational state upon substrate binding.
|
| |
Protein Sci,
15,
774-784.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Topalis,
B.Collinet,
C.Gasse,
L.Dugué,
J.Balzarini,
S.Pochet,
and
D.Deville-Bonne
(2005).
Substrate specificity of vaccinia virus thymidylate kinase.
|
| |
FEBS J,
272,
6254-6265.
|
 |
|
|
|
|
 |
M.Merdanovic,
E.Sauer,
and
J.Reidl
(2005).
Coupling of NAD+ biosynthesis and nicotinamide ribosyl transport: characterization of NadR ribonucleotide kinase mutants of Haemophilus influenzae.
|
| |
J Bacteriol,
187,
4410-4420.
|
 |
|
|
|
|
 |
C.Monnerjahn,
and
M.Konrad
(2003).
Modulated nucleoside kinases as tools to improve the activation of therapeutic nucleoside analogues.
|
| |
Chembiochem,
4,
143-146.
|
 |
|
|
|
|
 |
H.Munier-Lehmann,
A.Chaffotte,
S.Pochet,
and
G.Labesse
(2001).
Thymidylate kinase of Mycobacterium tuberculosis: a chimera sharing properties common to eukaryotic and bacterial enzymes.
|
| |
Protein Sci,
10,
1195-1205.
|
 |
|
|
|
|
 |
A.R.Van Rompay,
M.Johansson,
and
A.Karlsson
(2000).
Phosphorylation of nucleosides and nucleoside analogs by mammalian nucleoside monophosphate kinases.
|
| |
Pharmacol Ther,
87,
189-198.
|
 |
|
|
|
|
 |
I.Li de la Sierra,
H.Munier-Lehmann,
A.M.Gilles,
O.Bârzu,
and
M.Delarue
(2000).
Crystallization and preliminary X-ray analysis of the thymidylate kinase from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
226-228.
|
 |
|
|
|
|
 |
I.M.Li de La Sierra,
J.Gallay,
M.Vincent,
T.Bertrand,
P.Briozzo,
O.Bârzu,
and
A.M.Gilles
(2000).
Substrate-induced fit of the ATP binding site of cytidine monophosphate kinase from Escherichia coli: time-resolved fluorescence of 3'-anthraniloyl-2'-deoxy-ADP and molecular modeling.
|
| |
Biochemistry,
39,
15870-15878.
|
 |
|
|
|
|
 |
N.Ostermann,
I.Schlichting,
R.Brundiers,
M.Konrad,
J.Reinstein,
T.Veit,
R.S.Goody,
and
A.Lavie
(2000).
Insights into the phosphoryltransfer mechanism of human thymidylate kinase gained from crystal structures of enzyme complexes along the reaction coordinate.
|
| |
Structure,
8,
629-642.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Chenal-Francisque,
L.Tourneux,
E.Carniel,
P.Christova,
I.Li de la Sierra,
O.Bârzu,
and
A.M.Gilles
(1999).
The highly similar TMP kinases of Yersinia pestis and Escherichia coli differ markedly in their AZTMP phosphorylating activity.
|
| |
Eur J Biochem,
265,
112-119.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

