 |
PDBsum entry 4zst
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.1
- aryldialkylphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
An aryl dialkyl phosphate + H2O = dialkyl phosphate + an aryl alcohol
|
 |
 |
 |
 |
 |
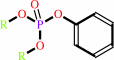 aryl dialkyl phosphate
aryl dialkyl phosphate
|
+
|
H2O
|
=
|
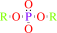 dialkyl phosphate
dialkyl phosphate
|
+
|
aryl alcohol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
54:5502-5512
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Variants of Phosphotriesterase for the Enhanced Detoxification of the Chemical Warfare Agent VR.
|
|
A.N.Bigley,
M.F.Mabanglo,
S.P.Harvey,
F.M.Raushel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The V-type organophosphorus nerve agents are among the most hazardous compounds
known. Previous efforts to evolve the bacterial enzyme phosphotriesterase (PTE)
for the hydrolytic decontamination of VX resulted in the identification of the
variant L7ep-3a, which has a kcat value more than 2 orders of magnitude higher
than that of wild-type PTE for the hydrolysis of VX. Because of the relatively
small size of the O-ethyl, methylphosphonate center in VX, stereoselectivity is
not a major concern. However, the Russian V-agent, VR, contains a larger
O-isobutyl, methylphosphonate center, making stereoselectivity a significant
issue since the SP-enantiomer is expected to be significantly more toxic than
the RP-enantiomer. The three-dimensional structure of the L7ep-3a variant was
determined to a resolution of 2.01 Å (PDB id: 4ZST ). The active site of the
L7ep-3a mutant has revealed a network of hydrogen bonding interactions between
Asp-301, Tyr-257, Gln-254, and the hydroxide that bridges the two metal ions. A
series of new analogues that mimic VX and VR has helped to identify critical
structural features for the development of new enzyme variants that are further
enhanced for the catalytic detoxification of VR and VX. The best of these
mutants has been shown to have a reversed stereochemical preference for the
hydrolysis of VR-chiral center analogues. This mutant hydrolyzes the two
enantiomers of VR 160- and 600-fold faster than wild-type PTE hydrolyzes the
SP-enantiomer of VR.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

