 |
PDBsum entry 4xby
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the l74f/m78v/i80v/l114f mutant of leh complexed with cyclopentene oxide
|
|
Structure:
|
 |
Limonene-1,2-epoxide hydrolase. Chain: c, d, a, b, e, f, g, h. Fragment: unp residues 5-149. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Rhodococcus erythropolis. Organism_taxid: 1833. Gene: lima. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.214
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
X.D.Kong,Z.Sun,J.H.Xu,M.T.Reetz,J.Zhou
|
|
Key ref:
|
 |
Z.Sun
et al.
(2015).
Reshaping an enzyme binding pocket for enhanced and inverted stereoselectivity: use of smallest amino acid alphabets in directed evolution.
Angew Chem Int Ed Engl,
54,
12410-12415.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
17-Dec-14
|
Release date:
|
15-Jul-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9ZAG3
(LIMA_RHOER) -
Limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
149 a.a.
144 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.3.2.8
- limonene-1,2-epoxide hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
limonene 1,2-epoxide + H2O = limonene-1,2-diol
|
 |
 |
 |
 |
 |
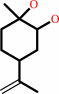
 limonene 1,2-epoxide
limonene 1,2-epoxide
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 54.55% similarity
|
=
|
 limonene-1,2-diol
limonene-1,2-diol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Angew Chem Int Ed Engl
54:12410-12415
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Reshaping an enzyme binding pocket for enhanced and inverted stereoselectivity: use of smallest amino acid alphabets in directed evolution.
|
|
Z.Sun,
R.Lonsdale,
X.D.Kong,
J.H.Xu,
J.Zhou,
M.T.Reetz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Directed evolution based on saturation mutagenesis at sites lining the binding
pocket is a commonly practiced strategy for enhancing or inverting the
stereoselectivity of enzymes for use in organic chemistry or biotechnology.
However, as the number of residues in a randomization site increases to five or
more, the screening effort for 95 % library coverage increases astronomically
until it is no longer feasible. We propose the use of a single amino acid for
saturation mutagenesis at superlarge randomization sites comprising 10 or more
residues. When used to reshape the binding pocket of limonene epoxide hydrolase,
this strategy, which drastically reduces the search space and thus the screening
effort, resulted in R,R- and S,S-selective mutants for the hydrolytic
desymmetrization of cyclohexene oxide and other epoxides. X-ray crystal
structures and docking studies of the mutants unveiled the source of
stereoselectivity and shed light on the mechanistic intricacies of this enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

