 |
PDBsum entry 4h6c
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of the allene oxide cyclase 1 from physcomitrella patens
|
|
Structure:
|
 |
Allene oxide cyclase. Chain: a, d, c, b, e, f, g, h, i, j, k, l. Engineered: yes
|
|
Source:
|
 |
Physcomitrella patens. Organism_taxid: 3218. Gene: aoc, aoc1. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.35Å
|
R-factor:
|
0.140
|
R-free:
|
0.174
|
|
|
Authors:
|
 |
P.Neumann,R.Ficner
|
|
Key ref:
|
 |
P.Neumann
et al.
(2012).
Crystal structures of Physcomitrella patens AOC1 and AOC2: insights into the enzyme mechanism and differences in substrate specificity.
Plant Physiol,
160,
1251-1266.
PubMed id:
|
 |
|
Date:
|
 |
|
19-Sep-12
|
Release date:
|
17-Oct-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8GS38
(Q8GS38_PHYPA) -
allene-oxide cyclase from Physcomitrium patens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
189 a.a.
180 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.99.6
- allene-oxide cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(9Z,13S,15Z)-12,13-epoxyoctadeca-9,11,15-trienoate = (9S,13S,15Z)-12- oxophyto-10,15-dienoate
|
 |
 |
 |
 |
 |
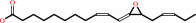 (9Z)-(13S)-12,13-epoxyoctadeca-9,11,15-trienoate
(9Z)-(13S)-12,13-epoxyoctadeca-9,11,15-trienoate
|
=
|
(15Z)-12-oxophyto- 10,15-dienoate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Plant Physiol
160:1251-1266
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of Physcomitrella patens AOC1 and AOC2: insights into the enzyme mechanism and differences in substrate specificity.
|
|
P.Neumann,
F.Brodhun,
K.Sauer,
C.Herrfurth,
M.Hamberg,
J.Brinkmann,
J.Scholz,
A.Dickmanns,
I.Feussner,
R.Ficner.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In plants, oxylipins regulate developmental processes and defense responses. The
first specific step in the biosynthesis of the cyclopentanone class of oxylipins
is catalyzed by allene oxide cyclase (AOC) that forms cis(+)-12-oxo-phytodienoic
acid. The moss Physcomitrella patens has two AOCs (PpAOC1 and PpAOC2) with
different substrate specificities for C(18)- and C(20)-derived substrates,
respectively. To better understand AOC's catalytic mechanism and to elucidate
the structural properties that explain the differences in substrate specificity,
we solved and analyzed the crystal structures of 36 monomers of both apo and
ligand complexes of PpAOC1 and PpAOC2. From these data, we propose the following
intermediates in AOC catalysis: (1) a resting state of the apo enzyme with a
closed conformation, (2) a first shallow binding mode, followed by (3) a tight
binding of the substrate accompanied by conformational changes in the binding
pocket, and (4) initiation of the catalytic cycle by opening of the epoxide
ring. As expected, the substrate dihydro analog
cis-12,13S-epoxy-9Z,15Z-octadecadienoic acid did not cyclize in the presence of
PpAOC1; however, when bound to the enzyme, it underwent isomerization into the
corresponding trans-epoxide. By comparing complex structures of the C(18)
substrate analog with in silico modeling of the C(20) substrate analog bound to
the enzyme allowed us to identify three major molecular determinants responsible
for the different substrate specificities (i.e. larger active site diameter, an
elongated cavity of PpAOC2, and two nonidentical residues at the entrance of the
active site).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

