 |
PDBsum entry 3q9c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of h159a apah complexed with n8-acetylspermidine
|
|
Structure:
|
 |
Acetylpolyamine amidohydrolase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Mycoplana ramosa. Mycoplana bullata. Organism_taxid: 40837. Gene: apha, aph. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.186
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
P.M.Lombardi,D.W.Christianson
|
|
Key ref:
|
 |
P.M.Lombardi
et al.
(2011).
Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases.
Biochemistry,
50,
1808-1817.
PubMed id:
|
 |
|
Date:
|
 |
|
07-Jan-11
|
Release date:
|
02-Mar-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q48935
(APAH_MYCRA) -
Acetylpolyamine amidohydrolase from Mycoplana ramosa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
341 a.a.
341 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.5.1.48
- acetylspermidine deacetylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N8-acetylspermidine + H2O = spermidine + acetate
|
 |
 |
 |
 |
 |
 N(8)-acetylspermidine
N(8)-acetylspermidine
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
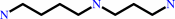 spermidine
spermidine
|
+
|
 acetate
acetate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.1.62
- acetylputrescine deacetylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-acetylputrescine + H2O = putrescine + acetate
|
 |
 |
 |
 |
 |
 N-acetylputrescine
N-acetylputrescine
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 69.23% similarity
|
=
|
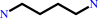 putrescine
putrescine
|
+
|
 acetate
acetate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
50:1808-1817
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases.
|
|
P.M.Lombardi,
H.D.Angell,
D.A.Whittington,
E.F.Flynn,
K.R.Rajashankar,
D.W.Christianson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

