 |
PDBsum entry 3o15
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of bacillus subtilis thiamin phosphate synthase complexed with a carboxylated thiazole phosphate
|
|
Structure:
|
 |
Thiamine-phosphate pyrophosphorylase. Chain: a. Synonym: tmp pyrophosphorylase, tmp-ppase, thiamine-phosphate synthase. Engineered: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Gene: thie. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.95Å
|
R-factor:
|
0.198
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
K.M.Mcculloch,J.W.Hanes,S.Abdelwahed,N.Mahanta,A.Hazra,K.Ishida, T.P.Begley,S.E.Ealick
|
|
Key ref:
|
 |
K.M.Mcculloch
et al.
Crystal structure and kinetic characterization of bac subtilis thiamin phosphate synthase with a carboxylat thiazole phosphate.
To be published,
.
|
 |
|
Date:
|
 |
|
20-Jul-10
|
Release date:
|
27-Jul-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P39594
(THIE_BACSU) -
Thiamine-phosphate synthase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
222 a.a.
219 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.3
- thiamine phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
2-[(2R,5Z)-2-carboxy-4-methylthiazol-5(2H)-ylidene]ethyl phosphate + 4-amino-2-methyl-5-(diphosphooxymethyl)pyrimidine + 2 H+ = thiamine phosphate + CO2 + diphosphate
|
|
2.
|
2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate + 4-amino-2-methyl- 5-(diphosphooxymethyl)pyrimidine + 2 H+ = thiamine phosphate + CO2 + diphosphate
|
|
3.
|
4-methyl-5-(2-phosphooxyethyl)-thiazole + 4-amino-2-methyl-5- (diphosphooxymethyl)pyrimidine + H+ = thiamine phosphate + diphosphate
|
|
 |
 |
 |
 |
 |
2-[(2R,5Z)-2-carboxy-4-methylthiazol-5(2H)-ylidene]ethyl phosphate
|
+
|
4-amino-2-methyl-5-(diphosphooxymethyl)pyrimidine
|
+
|
2
×
H(+)
|
=
|
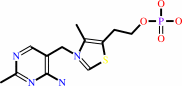 thiamine phosphate
thiamine phosphate
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
|
 |
 |
 |
 |
 |
2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate
|
+
|
4-amino-2-methyl- 5-(diphosphooxymethyl)pyrimidine
|
+
|
2
×
H(+)
|
=
|
 thiamine phosphate
thiamine phosphate
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
|
 |
 |
 |
 |
 |
4-methyl-5-(2-phosphooxyethyl)-thiazole
|
+
|
4-amino-2-methyl-5- (diphosphooxymethyl)pyrimidine
|
+
|
H(+)
|
=
|
thiamine phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

