 |
PDBsum entry 3nqb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of adenine deaminase from agrobacterium tumefaciens (str. C 58)
|
|
Structure:
|
 |
Adenine deaminase 2. Chain: a, b. Synonym: adenase 2, adenine aminase 2. Engineered: yes
|
|
Source:
|
 |
Agrobacterium tumefaciens. Organism_taxid: 176299. Strain: c58. Gene: ade2, adec, atu4426, agr_l_883. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.21Å
|
R-factor:
|
0.175
|
R-free:
|
0.235
|
|
|
Authors:
|
 |
A.Bagaria,D.Kumaran,S.K.Burley,S.Swaminathan,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
A.Bagaria
et al.
Crystal structure of adenine deaminase from agrobacte tumefaciens (str. C 58).
To be published,
.
|
 |
|
Date:
|
 |
|
29-Jun-10
|
Release date:
|
28-Jul-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q7CUX4
(ADEC2_AGRFC) -
Adenine deaminase 2 from Agrobacterium fabrum (strain C58 / ATCC 33970)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
597 a.a.
587 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.2
- adenine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
adenine + H2O + H+ = hypoxanthine + NH4+
|
 |
 |
 |
 |
 |
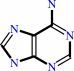 adenine
adenine
|
+
|
H2O
|
+
|
H(+)
|
=
|
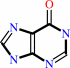 hypoxanthine
hypoxanthine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

