 |
PDBsum entry 3lvm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.8.1.7
- cysteine desulfurase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(sulfur carrier)-H + L-cysteine = (sulfur carrier)-SH + L-alanine
|
 |
 |
 |
 |
 |
(sulfur carrier)-H
|
+
|
 L-cysteine
L-cysteine
|
=
|
(sulfur carrier)-SH
|
+
|
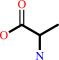 L-alanine
L-alanine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Plos Biol
8:e1000354
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
|
|
R.Shi,
A.Proteau,
M.Villarroya,
I.Moukadiri,
L.Zhang,
J.F.Trempe,
A.Matte,
M.E.Armengod,
M.Cygler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The cysteine desulfurase IscS is a highly conserved master enzyme initiating
sulfur transfer via persulfide to a range of acceptor proteins involved in Fe-S
cluster assembly, tRNA modifications, and sulfur-containing cofactor
biosynthesis. Several IscS-interacting partners including IscU, a scaffold for
Fe-S cluster assembly; TusA, the first member of a sulfur relay leading to
sulfur incorporation into the wobble uridine of several tRNAs; ThiI, involved in
tRNA modification and thiamine biosynthesis; and rhodanese RhdA are sulfur
acceptors. Other proteins, such as CyaY/frataxin and IscX, also bind to IscS,
but their functional roles are not directly related to sulfur transfer. We have
determined the crystal structures of IscS-IscU and IscS-TusA complexes providing
the first insight into their different modes of binding and the mechanism of
sulfur transfer. Exhaustive mutational analysis of the IscS surface allowed us
to map the binding sites of various partner proteins and to determine the
functional and biochemical role of selected IscS and TusA residues. IscS
interacts with its partners through an extensive surface area centered on the
active site Cys328. The structures indicate that the acceptor proteins approach
Cys328 from different directions and suggest that the conformational plasticity
of a long loop containing this cysteine is essential for the ability of IscS to
transfer sulfur to multiple acceptor proteins. The sulfur acceptors can only
bind to IscS one at a time, while frataxin and IscX can form a ternary complex
with IscU and IscS. Our data support the role of frataxin as an iron donor for
IscU to form the Fe-S clusters.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.D.Maynard,
D.N.Macklin,
K.Kirkegaard,
and
M.W.Covert
(2012).
Competing pathways control host resistance to virus via tRNA modification and programmed ribosomal frameshifting.
|
| |
Mol Syst Biol,
8,
567.
|
 |
|
|
|
|
 |
P.Subramanian,
A.V.Rodrigues,
S.Ghimire-Rijal,
and
T.L.Stemmler
(2011).
Iron chaperones for mitochondrial Fe-S cluster biosynthesis and ferritin iron storage.
|
| |
Curr Opin Chem Biol,
15,
312-318.
|
 |
|
|
|
|
 |
S.Rawat,
and
T.L.Stemmler
(2011).
Key players and their role during mitochondrial iron-sulfur cluster biosynthesis.
|
| |
Chemistry,
17,
746-753.
|
 |
|
|
|
|
 |
S.Schmucker,
A.Martelli,
F.Colin,
A.Page,
M.Wattenhofer-Donzé,
L.Reutenauer,
and
H.Puccio
(2011).
Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex.
|
| |
PLoS One,
6,
e16199.
|
 |
|
|
|
|
 |
B.Py,
and
F.Barras
(2010).
Building Fe-S proteins: bacterial strategies.
|
| |
Nat Rev Microbiol,
8,
436-446.
|
 |
|
|
|
|
 |
F.Prischi,
P.V.Konarev,
C.Iannuzzi,
C.Pastore,
S.Adinolfi,
S.R.Martin,
D.I.Svergun,
and
A.Pastore
(2010).
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
|
| |
Nat Commun,
1,
95.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

