 |
PDBsum entry 3hxx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.7
- alanine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Ala) + L-alanine + ATP = L-alanyl-tRNA(Ala) + AMP + diphosphate
|
 |
 |
 |
 |
 |
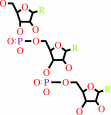 tRNA(Ala)
tRNA(Ala)
|
+
|
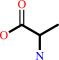 L-alanine
L-alanine
|
+
|
 ATP
ATP
|
=
|
L-alanyl-tRNA(Ala)
Bound ligand (Het Group name = )
matches with 74.19% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nature
462:808-812
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
|
|
M.Guo,
Y.E.Chong,
R.Shapiro,
K.Beebe,
X.L.Yang,
P.Schimmel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mistranslation arising from confusion of serine for alanine by alanyl-tRNA
synthetases (AlaRSs) has profound functional consequences. Throughout evolution,
two editing checkpoints prevent disease-causing mistranslation from confusing
glycine or serine for alanine at the active site of AlaRS. In both bacteria and
mice, Ser poses a bigger challenge than Gly. One checkpoint is the AlaRS editing
centre, and the other is from widely distributed AlaXps-free-standing,
genome-encoded editing proteins that clear Ser-tRNA(Ala). The paradox of
misincorporating both a smaller (glycine) and a larger (serine) amino acid
suggests a deep conflict for nature-designed AlaRS. Here we show the chemical
basis for this conflict. Nine crystal structures, together with kinetic and
mutational analysis, provided snapshots of adenylate formation for each amino
acid. An inherent dilemma is posed by constraints of a structural design that
pins down the alpha-amino group of the bound amino acid by using an acidic
residue. This design, dating back more than 3 billion years, creates a
serendipitous interaction with the serine OH that is difficult to avoid.
Apparently because no better architecture for the recognition of alanine could
be found, the serine misactivation problem was solved through free-standing
AlaXps, which appeared contemporaneously with early AlaRSs. The results reveal
unconventional problems and solutions arising from the historical design of the
protein synthesis machinery.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.H.Nawaz,
E.Merriman,
X.L.Yang,
and
P.Schimmel
(2011).
p23H implicated as cis/trans regulator of AlaXp-directed editing for mammalian cell homeostasis.
|
| |
Proc Natl Acad Sci U S A,
108,
2723-2728.
|
 |
|
|
|
|
 |
M.Guo,
R.Shapiro,
P.Schimmel,
and
X.L.Yang
(2010).
Introduction of a leucine half-zipper engenders multiple high-quality crystals of a recalcitrant tRNA synthetase.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
243-250.
|
 |
|
|
|
|
 |
N.M.Reynolds,
B.A.Lazazzera,
and
M.Ibba
(2010).
Cellular mechanisms that control mistranslation.
|
| |
Nat Rev Microbiol,
8,
849-856.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

