 |
PDBsum entry 3f1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of 2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase from listeria monocytogenes
|
|
Structure:
|
 |
Putative 2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase 2. Chain: a, b. Synonym: 4-diphosphocytidyl-2c-methyl-d-erythritol synthase 2, mep cytidylyltransferase 2, mct 2. Engineered: yes
|
|
Source:
|
 |
Listeria monocytogenes str. 4b f2365. Organism_taxid: 265669. Gene: ispd2, lmof2365_1100. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.242
|
R-free:
|
0.277
|
|
|
Authors:
|
 |
Y.Patskovsky,J.Ho,R.Toro,M.Gilmore,S.Miller,C.Groshong,J.M.Sauder, S.K.Burley,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
Y.Patskovsky
et al.
Crystal structure of 2-C-Methyl-D-Erythritol 4-Phosphate cytidylyltransferase from listeria monocytogenes.
To be published,
.
|
 |
|
Date:
|
 |
|
27-Oct-08
|
Release date:
|
18-Nov-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q720Y7
(TARI_LISMF) -
Ribitol-5-phosphate cytidylyltransferase from Listeria monocytogenes serotype 4b (strain F2365)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
237 a.a.
229 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.40
- D-ribitol-5-phosphate cytidylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-ribitol 5-phosphate + CTP + H+ = CDP-L-ribitol + diphosphate
|
 |
 |
 |
 |
 |
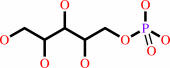 D-ribitol 5-phosphate
D-ribitol 5-phosphate
|
+
|
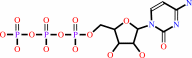 CTP
CTP
|
+
|
H(+)
|
=
|
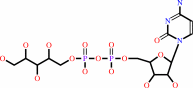 CDP-L-ribitol
CDP-L-ribitol
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

