 |
PDBsum entry 3etl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
DNA binding protein, recombination
|
PDB id
|
|

|

|
3etl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
65:602-610
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
|
|
Y.Li,
Y.He,
Y.Luo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Archaeal RadAs are close homologues of eukaryal Rad51s ( approximately 40%
sequence identity). These recombinases promote ATP hydrolysis and a hallmark
strand-exchange reaction between homologous single-stranded and double-stranded
DNA substrates. Pairing of the 3'-overhangs located at the damaged DNA with a
homologous double-stranded DNA enables the re-synthesis of the damaged region
using the homologous DNA as the template. In recent studies, conformational
changes in the DNA-interacting regions of Methanococcus voltae RadA have been
correlated with the presence of activity-stimulating potassium or calcium ions
in the ATPase centre. The series of crystal structures of M. maripaludis RadA
presented here further suggest the conservation of an allosteric switch in the
ATPase centre which controls the conformational status of DNA-interacting loops.
Structural comparison with the distant Escherichia coli RecA homologue supports
the notion that the conserved Lys248 and Lys250 residues in RecA play a role
similar to that of cations in RadA. The conservation of a cationic bridge
between the DNA-interacting L2 region and the terminal phosphate of ATP,
together with the apparent stability of the nucleoprotein filament, suggests a
gap-displacement model which may explain the advantage of ATP hydrolysis for
DNA-strand exchange.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4 Superimposed ATPase centres of MmRadA and EcRecA in
stereo. The conserved P-loops are superimposed. The P-loop
(Gly105-Thr112), His280 and Asp302 of the ATPase-active form of
MmRadA are shown as green stick models. The MmRadA-bound AMPPNP
and monovalent cations are shown as yellow ball-and-stick
models. The P-loop (Gly66-Thr73), Phe217, Lys248 and Lys250 of
the active EcRecA structure are shown in blue.
|
 |
Figure 5.
Figure 5 Hypothetical model of ATP hydrolysis-facilitated gap
displacement. The crystallized protein filament is shown as a C^
 trace
in salmon with AMPPNP in green. Speculative models of the DNA
substrates are shown as wires. Important segments are boxed. The
5'- and 3'-ends are based on the filament-initiating ssDNA
(thinner wire in blue). The homologous dsDNA is shown as a
thicker wire in yellow. The strand-exchange process progresses
from the 5'-end to the 3'-end. (a) An intervening gap. Such gaps
are likely to exist owing to simultaneous homologous pairing
between the recombinase/ssDNA filament and dsDNA at multiple
locations. The dsDNA in the gap region cannot become properly
wound ( trace
in salmon with AMPPNP in green. Speculative models of the DNA
substrates are shown as wires. Important segments are boxed. The
5'- and 3'-ends are based on the filament-initiating ssDNA
(thinner wire in blue). The homologous dsDNA is shown as a
thicker wire in yellow. The strand-exchange process progresses
from the 5'-end to the 3'-end. (a) An intervening gap. Such gaps
are likely to exist owing to simultaneous homologous pairing
between the recombinase/ssDNA filament and dsDNA at multiple
locations. The dsDNA in the gap region cannot become properly
wound ( 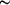 19
bp per helical turn) around the nucleoprotein filament without
unwinding its adjacent region(s). Despite the sequence homology,
it serves as a topological roadblock of strand exchange between
long DNA substrates. (b) ATP hydrolysis promotes the transient
release of a dsDNA segment at the immediate 3'-flank of the gap.
The transiently released dsDNA region is shown as an exaggerated
wide helix. (c) Rearrangement in the transiently released dsDNA
region and the adjacent gap takes place without changing the
overall topology. The 5'-end of the gap region becomes properly
wound, while the released dsDNA region becomes unwound. (d) The
rearranged 5'-end of the gap becomes bound by the recombinase
filament. As a result, the gap is displaced towards the 3'-end.
(e) Repetition of steps (b)-(d) would chase the topologically
strained gap out of the 3'-end of the nucleoprotein filament,
therefore removing topological roadblocks to extensive DNA
strand exchange. 19
bp per helical turn) around the nucleoprotein filament without
unwinding its adjacent region(s). Despite the sequence homology,
it serves as a topological roadblock of strand exchange between
long DNA substrates. (b) ATP hydrolysis promotes the transient
release of a dsDNA segment at the immediate 3'-flank of the gap.
The transiently released dsDNA region is shown as an exaggerated
wide helix. (c) Rearrangement in the transiently released dsDNA
region and the adjacent gap takes place without changing the
overall topology. The 5'-end of the gap region becomes properly
wound, while the released dsDNA region becomes unwound. (d) The
rearranged 5'-end of the gap becomes bound by the recombinase
filament. As a result, the gap is displaced towards the 3'-end.
(e) Repetition of steps (b)-(d) would chase the topologically
strained gap out of the 3'-end of the nucleoprotein filament,
therefore removing topological roadblocks to extensive DNA
strand exchange.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the IUCr:
Acta Crystallogr D Biol Crystallogr
(2009,
65,
602-610)
copyright 2009.
|
|
| |
Figures were
selected
by the author.
|
|

|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
The apparent conservation of an ATPase switch which alternates the DNA-binding loops between high-affinity and low-affinity states suggests a gap displacement model which would enable the optimal justaposition of single and double-stranded DNA substrates.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

