 |
PDBsum entry 3dbm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.92
- hydroperoxide dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(13S)-hydroperoxy-(9Z,11E,15Z)-octadecatrienoate = (9Z,13S,15Z)-12,13- epoxyoctadeca-9,11,15-trienoate + H2O
|
 |
 |
 |
 |
 |
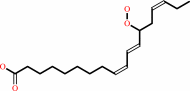 (9Z,11E,15Z)-(13S)-hydroperoxyoctadeca-9,11,15-trienoate
(9Z,11E,15Z)-(13S)-hydroperoxyoctadeca-9,11,15-trienoate
|
=
|
(9Z,15Z)- (13S)-12,13-epoxyoctadeca-9,11,15-trienoate
|
+
|
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme-thiolate
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
105:13883-13888
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
|
|
L.Li,
Z.Chang,
Z.Pan,
Z.Q.Fu,
X.Wang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cytochrome P450s exist ubiquitously in all organisms and are involved in many
biological processes. Allene oxide synthase (AOS) is a P450 enzyme that plays a
key role in the biosynthesis of oxylipin jasmonates, which are involved in
signal and defense reactions in higher plants. The crystal structures of guayule
(Parthenium argentatum) AOS (CYP74A2) and its complex with the substrate analog
13(S)-hydroxyoctadeca-9Z,11E-dienoic acid have been determined. The structures
exhibit a classic P450 fold but possess a heme-binding mode with an unusually
long heme binding loop and a unique I-helix. The structures also reveal two
channels through which substrate and product may access and leave the active
site. The entrances are defined by a loop between beta3-2 and beta3-3. Asn-276
in the substrate binding site may interact with the substrate's hydroperoxy
group and play an important role in catalysis, and Lys-282 at the entrance may
control substrate access and binding. These studies provide both structural
insights into AOS and related P450s and a structural basis to understand the
distinct reaction mechanism.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Ribbon diagram of the structure of AOS with bound heme and
13(S)-HODE. The α- and β-domains are shown in cyan and magenta
with the secondary structures and the N and C termini labeled.
The heme and 13(S)-HODE molecules are shown as ball-and-stick
models. Figs. 2, 3, 4A, and 5 were prepared with MOLSCRIPT (45)
and RASTER3D (46).
|
 |
Figure 3.
Heme-binding site. (A) Stereo diagram shows heme molecule and
its interactions with AOS. The structure of heme is shown as a
ball-and-stick model. (B) Stereo diagram shows a comparison of
I-helix of AOS (in cyan) and human P450 2C9 (in gray). (C)
Stereo diagram shows a comparison of heme-binding loop of AOS
(in cyan) and human P450 2C9 (in gray).
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Nelson,
and
D.Werck-Reichhart
(2011).
A P450-centric view of plant evolution.
|
| |
Plant J,
66,
194-211.
|
 |
|
|
|
|
 |
X.Wang
(2011).
Structure, function, and engineering of enzymes in isoflavonoid biosynthesis.
|
| |
Funct Integr Genomics,
11,
13-22.
|
 |
|
|
|
|
 |
M.Mizutani,
and
D.Ohta
(2010).
Diversification of P450 genes during land plant evolution.
|
| |
Annu Rev Plant Biol,
61,
291-315.
|
 |
|
|
|
|
 |
T.C.Pochapsky,
S.Kazanis,
and
M.Dang
(2010).
Conformational plasticity and structure/function relationships in cytochromes P450.
|
| |
Antioxid Redox Signal,
13,
1273-1296.
|
 |
|
|
|
|
 |
A.R.Brash
(2009).
Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes.
|
| |
Phytochemistry,
70,
1522-1531.
|
 |
|
|
|
|
 |
I.R.Chechetkin,
F.K.Mukhitova,
A.S.Blufard,
A.Y.Yarin,
L.L.Antsygina,
and
A.N.Grechkin
(2009).
Unprecedented pathogen-inducible complex oxylipins from flax--linolipins A and B.
|
| |
FEBS J,
276,
4463-4472.
|
 |
|
|
|
|
 |
R.Hänsch,
and
R.R.Mendel
(2009).
Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl).
|
| |
Curr Opin Plant Biol,
12,
259-266.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

