 |
PDBsum entry 3d2c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structure of 4d3, a thermostable mutant of bacillus subtilis lipase obtained through directed evolution
|
|
Structure:
|
 |
Lipase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Synonym: triacylglycerol lipase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Gene: lipa, lip, bsu02700. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.18Å
|
R-factor:
|
0.202
|
R-free:
|
0.245
|
|
|
Authors:
|
 |
R.Sankaranarayanan,M.Z.Kamal
|
Key ref:

|
 |
S.Ahmad
et al.
(2008).
Thermostable Bacillus subtilis lipases: in vitro evolution and structural insight.
J Mol Biol,
381,
324-340.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-May-08
|
Release date:
|
03-Jun-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P37957
(ESTA_BACSU) -
Lipase EstA from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
212 a.a.
179 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 9 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.3
- triacylglycerol lipase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a triacylglycerol + H2O = a diacylglycerol + a fatty acid + H+
|
 |
 |
 |
 |
 |
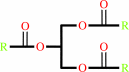 triacylglycerol
triacylglycerol
|
+
|
H2O
|
=
|
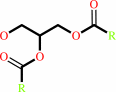 diacylglycerol
diacylglycerol
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
381:324-340
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Thermostable Bacillus subtilis lipases: in vitro evolution and structural insight.
|
|
S.Ahmad,
M.Z.Kamal,
R.Sankaranarayanan,
N.M.Rao.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In vitro evolution methods are now being routinely used to identify protein
variants with novel and enhanced properties that are difficult to achieve using
rational design. However, one of the limitations is in screening for beneficial
mutants through several generations due to the occurrence of neutral/negative
mutations occurring in the background of positive ones. While evolving a lipase
in vitro from mesophilic Bacillus subtilis to generate thermostable variants, we
have designed protocols that combine stringent three-tier testing, sequencing
and stability assessments on the protein at the end of each generation. This
strategy resulted in a total of six stabilizing mutations in just two
generations with three mutations per generation. Each of the six mutants when
evaluated individually contributed additively to thermostability. A combination
of all of them resulted in the best variant that shows a remarkable 15 degrees C
shift in melting temperature and a millionfold decrease in the thermal
inactivation rate with only a marginal increase of 3 kcal mol(-1) in free energy
of stabilization. Notably, in addition to the dramatic shift in optimum
temperature by 20 degrees C, the activity has increased two- to fivefold in the
temperature range 25-65 degrees C. High-resolution crystal structures of three
of the mutants, each with 5 degrees increments in melting temperature, reveal
the structural basis of these mutations in attaining higher thermostability. The
structures highlight the importance of water-mediated ionic networks on the
protein surface in imparting thermostability. Saturation mutagenesis at each of
the six positions did not result in enhanced thermostability in almost all the
cases, confirming the crucial role played by each mutation as revealed through
the structural study. Overall, our study presents an efficient strategy that can
be employed in directed evolution approaches employed for obtaining improved
properties of proteins.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Fig. 5. Ribbon diagram of the LipA molecule indicating the
relative position of each mutation.
|
 |
Figure 7.
Fig. 7. Structural changes in the regions around mutations
identified in the third generation. (a) A15S mutation in the
background of F17S mutation makes a hydrogen bond with the same.
(b) Region around mutation A20E showing a water molecule making
hydrogen bonds with side chains of residues Glu20, Ser24 and
Arg33. (c) Electrostatic network consisting of side chains of
Arg107 and Asp144, main-chain carbonyl groups of residues 107
and 142 and three water molecules named WT1, WT2 and WT3 in the
wild-type protein. (d) Immediate vicinity around G111D mutation
showing WT2 is replaced by carboxylate oxygen of Asp111 in the
mutant. Stick representations of loop regions 107–108 and
142–143 are overlaid on the ribbon representations. In all the
figures, water molecules are shown as red spheres and small blue
spheres show electrostatic interactions.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
381,
324-340)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Chakravorty,
S.Parameswaran,
V.K.Dubey,
and
S.Patra
(2011).
In silico characterization of thermostable lipases.
|
| |
Extremophiles,
15,
89.
|
 |
|
|
|
|
 |
J.Khurana,
R.Singh,
and
J.Kaur
(2011).
Engineering of Bacillus lipase by directed evolution for enhanced thermal stability: effect of isoleucine to threonine mutation at protein surface.
|
| |
Mol Biol Rep,
38,
2919-2926.
|
 |
|
|
|
|
 |
D.Y.Colin,
P.Deprez-Beauclair,
N.Silva,
L.Infantes,
and
B.Kerfelec
(2010).
Modification of pancreatic lipase properties by directed molecular evolution.
|
| |
Protein Eng Des Sel,
23,
365-373.
|
 |
|
|
|
|
 |
S.Ahmad,
and
N.M.Rao
(2009).
Thermally denatured state determines refolding in lipase: mutational analysis.
|
| |
Protein Sci,
18,
1183-1196.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

