 |
PDBsum entry 3a2k

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.19
- tRNA(Ile)-lysidine synthetase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cytidine34 in tRNA(Ile2) + L-lysine + ATP = lysidine34 in tRNA(Ile2) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
cytidine(34) in tRNA(Ile2)
|
+
|
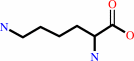 L-lysine
L-lysine
|
+
|
 ATP
ATP
|
=
|
lysidine(34) in tRNA(Ile2)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
461:1144-1148
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
|
|
K.Nakanishi,
L.Bonnefond,
S.Kimura,
T.Suzuki,
R.Ishitani,
O.Nureki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Maturation of precursor transfer RNA (pre-tRNA) includes excision of the 5'
leader and 3' trailer sequences, removal of introns and addition of the CCA
terminus. Nucleotide modifications are incorporated at different stages of tRNA
processing, after the RNA molecule adopts the proper conformation. In bacteria,
tRNA(Ile2) lysidine synthetase (TilS) modifies cytidine into lysidine (L;
2-lysyl-cytidine) at the first anticodon of tRNA(Ile2) (refs 4-9). This
modification switches tRNA(Ile2) from a methionine-specific to an
isoleucine-specific tRNA. However, the aminoacylation of tRNA(Ile2) by
methionyl-tRNA synthetase (MetRS), before the modification by TilS, might lead
to the misincorporation of methionine in response to isoleucine codons. The
mechanism used by bacteria to avoid this pitfall is unknown. Here we show that
the TilS enzyme specifically recognizes and modifies tRNA(Ile2) in its precursor
form, thereby avoiding translation errors. We identified the lysidine
modification in pre-tRNA(Ile2) isolated from RNase-E-deficient Escherichia coli
and did not detect mature tRNA(Ile2) lacking this modification. Our kinetic
analyses revealed that TilS can modify both types of RNA molecule with
comparable efficiencies. X-ray crystallography and mutational analyses revealed
that TilS specifically recognizes the entire L-shape structure in pre-tRNA(Ile2)
through extensive interactions coupled with sequential domain movements. Our
results demonstrate how TilS prevents the recognition of tRNA(Ile2) by MetRS and
achieves high specificity for its substrate. These two key points form the basis
for maintaining the fidelity of isoleucine codon translation in bacteria. Our
findings also provide a rationale for the necessity of incorporating specific
modifications at the precursor level during tRNA biogenesis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2: tRNA recognition by GkTilS. a, Overall homodimeric
structure. b, Recognition of the anticodon loop. F[o ]- F[c]
simulated annealing omit maps (3.7  )
of Arg 142 and three nucleotides (C32, C34 and A38) are shown in
magenta and grey, respectively. c, Left, recognition of the
major groove of the acceptor stem by the HTH motif and )
of Arg 142 and three nucleotides (C32, C34 and A38) are shown in
magenta and grey, respectively. c, Left, recognition of the
major groove of the acceptor stem by the HTH motif and  -hairpin;
right, the protein surface that complementarily interacts with
the 3'-ACCA terminus is coloured according to its electrostatic
potential. d, Lysidine incorporating activities of tRNA^Ile2 and
GkTilS mutants. The initial rate of lysidine incorporation is
shown. Error bars, s.d. of three independent experiments. -hairpin;
right, the protein surface that complementarily interacts with
the 3'-ACCA terminus is coloured according to its electrostatic
potential. d, Lysidine incorporating activities of tRNA^Ile2 and
GkTilS mutants. The initial rate of lysidine incorporation is
shown. Error bars, s.d. of three independent experiments.
|
 |
Figure 3.
Figure 3: Sequential tRNA recognition mechanism. a,
Superimposition of apo-EcTilS and tRNA-bound GkTilS on their
catalytic domains. b, Superimposition of type I and II TilSs on
their SCL domains. c, Lysidine incorporation into tRNA^Mets by
GkTilS (left) and AaTilS (right). Error bars denote s.d. of
three independent experiments. d, Lysidine formation model. e,
Apo-form (apo-EcTilS): the ASB and SCL domains have an
intramolecular hydrophobic interaction. f, Initial binding state
(manual docking of yeast tRNA^Phe (PDB accession 1EHZ) onto the
apo-EcTilS): capture of the pre-tRNA^Ile2 acceptor stem by the
ASB domain triggers disruption of the hydrophobic interactions.
g, Pre-reaction state (the current structure): drastic domain
movements allow TilS to interact fully with tRNA.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(2009,
461,
1144-1148)
copyright 2009.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Guelorget,
and
B.Golinelli-Pimpaneau
(2011).
Mechanism-based strategies for trapping and crystallizing complexes of RNA-modifying enzymes.
|
| |
Structure,
19,
282-291.
|
 |
|
|
|
|
 |
C.Fabret,
E.Dervyn,
B.Dalmais,
A.Guillot,
C.Marck,
H.Grosjean,
and
P.Noirot
(2011).
Life without the essential bacterial tRNA(Ile2) -lysidine synthetase TilS: a case of tRNA gene recruitment in Bacillus subtilis.
|
| |
Mol Microbiol,
80,
1062-1074.
|
 |
|
|
|
|
 |
N.Terasaka,
S.Kimura,
T.Osawa,
T.Numata,
and
T.Suzuki
(2011).
Biogenesis of 2-agmatinylcytidine catalyzed by the dual protein and RNA kinase TiaS.
|
| |
Nat Struct Mol Biol,
18,
1268-1274.
|
 |
|
|
|
|
 |
R.L.Sherrer,
Y.Araiso,
C.Aldag,
R.Ishitani,
J.M.Ho,
D.Söll,
and
O.Nureki
(2011).
C-terminal domain of archaeal O-phosphoseryl-tRNA kinase displays large-scale motion to bind the 7-bp D-stem of archaeal tRNA(Sec).
|
| |
Nucleic Acids Res,
39,
1034-1041.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Osawa,
S.Kimura,
N.Terasaka,
H.Inanaga,
T.Suzuki,
and
T.Numata
(2011).
Structural basis of tRNA agmatinylation essential for AUA codon decoding.
|
| |
Nat Struct Mol Biol,
18,
1275-1280.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Kawai,
and
S.Yokoyama
(2010).
Professor Tatsuo Miyazawa: from molecular structure to biological function.
|
| |
J Biochem,
148,
631-638.
|
 |
|
|
|
|
 |
Y.Ikeuchi,
S.Kimura,
T.Numata,
D.Nakamura,
T.Yokogawa,
T.Ogata,
T.Wada,
T.Suzuki,
and
T.Suzuki
(2010).
Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea.
|
| |
Nat Chem Biol,
6,
277-282.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

