 |
PDBsum entry 3cy6

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Unknown function
|
PDB id
|
|

|

|
3cy6
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.124
- protein deglycase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N(omega)-(1-hydroxy-2-oxopropyl)-L-arginyl-[protein] + H2O = lactate + L-arginyl-[protein] + H+
|
|
2.
|
N6-(1-hydroxy-2-oxopropyl)-L-lysyl-[protein] + H2O = lactate + L-lysyl-[protein] + H+
|
|
3.
|
S-(1-hydroxy-2-oxopropyl)-L-cysteinyl-[protein] + H2O = lactate + L-cysteinyl-[protein] + H+
|
|
 |
 |
 |
 |
 |
N(omega)-(1-hydroxy-2-oxopropyl)-L-arginyl-[protein]
|
+
|
H2O
|
=
|
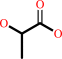 lactate
lactate
|
+
|
L-arginyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
N(6)-(1-hydroxy-2-oxopropyl)-L-lysyl-[protein]
|
+
|
H2O
|
=
|
 lactate
lactate
|
+
|
L-lysyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
S-(1-hydroxy-2-oxopropyl)-L-cysteinyl-[protein]
|
+
|
H2O
|
=
|
 lactate
lactate
|
+
|
L-cysteinyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:7430-7440
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
|
|
A.C.Witt,
M.Lakshminarasimhan,
B.C.Remington,
S.Hasim,
E.Pozharski,
M.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human DJ-1, a disease-associated protein that protects cells from oxidative
stress, contains an oxidation-sensitive cysteine (C106) that is essential for
its cytoprotective activity. The origin of C106 reactivity is obscure, due in
part to the absence of an experimentally determined p K a value for this
residue. We have used atomic-resolution X-ray crystallography and UV
spectroscopy to show that C106 has a depressed p K a of 5.4 +/- 0.1 and that the
C106 thiolate accepts a hydrogen bond from a protonated glutamic acid side chain
(E18). X-ray crystal structures and cysteine p K a analysis of several
site-directed substitutions at residue 18 demonstrate that the protonated
carboxylic acid side chain of E18 is required for the maximal stabilization of
the C106 thiolate. A nearby arginine residue (R48) participates in a guanidinium
stacking interaction with R28 from the other monomer in the DJ-1 dimer and
elevates the p K a of C106 by binding an anion that electrostatically suppresses
thiol ionization. Our results show that the ionizable residues (E18, R48, and
R28) surrounding C106 affect its p K a in a way that is contrary to expectations
based on the typical ionization behavior of glutamic acid and arginine. Lastly,
a search of the Protein Data Bank (PDB) produces several candidate
hydrogen-bonded aspartic/glutamic acid-cysteine interactions, which we propose
are particularly common in the DJ-1 superfamily.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Blackinton,
M.Lakshminarasimhan,
K.J.Thomas,
R.Ahmad,
E.Greggio,
A.S.Raza,
M.R.Cookson,
and
M.A.Wilson
(2009).
Formation of a Stabilized Cysteine Sulfinic Acid Is Critical for the Mitochondrial Function of the Parkinsonism Protein DJ-1.
|
| |
J Biol Chem,
284,
6476-6485.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Waak,
S.S.Weber,
K.Görner,
C.Schall,
H.Ichijo,
T.Stehle,
and
P.J.Kahle
(2009).
Oxidizable Residues Mediating Protein Stability and Cytoprotective Interaction of DJ-1 with Apoptosis Signal-regulating Kinase 1.
|
| |
J Biol Chem,
284,
14245-14257.
|
 |
|
|
|
|
 |
P.Acín,
J.Rayó,
A.Guerrero,
and
C.Quero
(2009).
Improved resolution in the acidic and basic region of 2-DE of insect antennae proteins using hydroxyethyl disulfide.
|
| |
Electrophoresis,
30,
2613-2616.
|
 |
|
|
|
|
 |
P.J.Kahle,
J.Waak,
and
T.Gasser
(2009).
DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders.
|
| |
Free Radic Biol Med,
47,
1354-1361.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

