 |
PDBsum entry 2z3h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of blasticidin s deaminase (bsd) complexed with deaminohydroxy blasticidin s
|
|
Structure:
|
 |
Blasticidin-s deaminase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Aspergillus terreus. Organism_taxid: 33178. Strain: s-712. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.173
|
R-free:
|
0.185
|
|
|
Authors:
|
 |
T.Kumasaka,M.Yamamoto,M.Furuichi,M.Nakasako,M.Kimura,I.Yamaguchi, T.Ueki
|
Key ref:

|
 |
T.Kumasaka
et al.
(2007).
Crystal Structures of Blasticidin S Deaminase (BSD): IMPLICATIONS FOR DYNAMIC PROPERTIES OF CATALYTIC ZINC.
J Biol Chem,
282,
37103-37111.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
04-Jun-07
|
Release date:
|
23-Oct-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0C2P0
(BSD_ASPTE) -
Blasticidin-S deaminase from Aspergillus terreus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
130 a.a.
122 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.23
- blasticidin-S deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
blasticidin S + H2O + H+ = deaminohydroxyblasticidin S + NH4+
|
 |
 |
 |
 |
 |
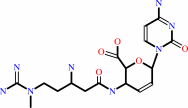 blasticidin S
blasticidin S
|
+
|
H2O
|
+
|
H(+)
|
=
|
deaminohydroxyblasticidin S
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:37103-37111
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal Structures of Blasticidin S Deaminase (BSD): IMPLICATIONS FOR DYNAMIC PROPERTIES OF CATALYTIC ZINC.
|
|
T.Kumasaka,
M.Yamamoto,
M.Furuichi,
M.Nakasako,
A.H.Teh,
M.Kimura,
I.Yamaguchi,
T.Ueki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The set of blasticidin S (BS) and blasticidin S deaminase (BSD) is a widely used
selectable marker for gene transfer experiments. BSD is a member of the cytidine
deaminase (CDA) family; it is a zinc-dependent enzyme with three cysteines and
one water molecule as zinc ligands. The crystal structures of BSD were
determined in six states (i.e. native, substrate-bound, product-bound,
cacodylate-bound, substrate-bound E56Q mutant, and R90K mutant). In the
structures, the zinc position and coordination structures vary. The
substrate-bound structure shows a large positional and geometrical shift of zinc
with a double-headed electron density of the substrate that seems to be assigned
to the amino and hydroxyl groups of the substrate and product, respectively. In
this intermediate-like structure, the steric hindrance of the hydroxyl group
pushes the zinc into the triangular plane consisting of three cysteines with a
positional shift of approximately 0.6 A, and the fifth ligand water approaches
the opposite direction of the substrate with a shift of 0.4 A(.) Accordingly,
the zinc coordination is changed from tetrahedral to trigonal bipyramidal, and
its coordination distance is extended between zinc and its intermediate. The
shift of zinc and the recruited water is also observed in the structure of the
inactivated E56Q mutant. This novel observation is different in two-cysteine
cytidine deaminase Escherichia coli CDA and might be essential for the reaction
mechanism in BSD, since it is useful for the easy release of the product by
charge compensation and for the structural change of the substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Structural comparison of catalytic center and tetrameric
interface.A, BSD; B, BSD R90K mutant; C, EcCDA; D, BsCDA; E,
MmCDA. D and E show that 16 and 18 water molecules occupied the
region, respectively. The central cavities are drawn as gray
surfaces. There is no cavity in BSD or its mutant.
|
 |
Figure 4.
Tetrahedral and trigonal bipyramidal coordinations of zinc
with omit F[O] - F[C] maps.A, native; B, B-subunit of substrate
complex. Number sign (#) indicates Ser^86 carbonyl oxygen. C,
A-subunit of substrate complex. All the omit F[O] - F[C] maps
contoured at the 4.0σ level. Five omitted groups are shown in
different colors: zinc (yellow); Cys^54, Cys^88, and Cys^91
(orange); Glu^56 (magenta); Arg^90 (lime); fourth and fifth
ligands (cyan). Only in the calculation of fourth and fifth
ligands were an additional twenty refinement cycles by Refmac
performed to eliminate model bias.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
37103-37111)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

