 |
PDBsum entry 2uyu

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.19
- rhamnulose-1-phosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-rhamnulose 1-phosphate = (S)-lactaldehyde + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
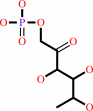 L-rhamnulose 1-phosphate
L-rhamnulose 1-phosphate
|
=
|
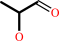 (S)-lactaldehyde
(S)-lactaldehyde
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
319:206-209
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Designed protein-protein association.
|
|
D.Grueninger,
N.Treiber,
M.O.Ziegler,
J.W.Koetter,
M.S.Schulze,
G.E.Schulz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The analysis of natural contact interfaces between protein subunits and between
proteins has disclosed some general rules governing their association. We have
applied these rules to produce a number of novel assemblies, demonstrating that
a given protein can be engineered to form contacts at various points of its
surface. Symmetry plays an important role because it defines the multiplicity of
a designed contact and therefore the number of required mutations. Some of the
proteins needed only a single side-chain alteration in order to associate to a
higher-order complex. The mobility of the buried side chains has to be taken
into account. Four assemblies have been structurally elucidated. Comparisons
between the designed contacts and the results will provide useful guidelines for
the development of future architectures.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Design of protein assemblies (24). The proteins in (C)
to (H) are depicted as thick-lined C  plots at various
scales with mutated residues as colored spheres. (A) Sketch of
an asymmetric interface between patches a and b, which, in
general, gives rise to an infinite helix (top). A C[2]-symmetric
interface also between patches a and b doubles the numbers of
contacts and forms a globular complex (bottom). Along the same
lines, the reported D[2], D[4], and D[8] oligomers have 4-, 8-,
and 16-fold contacts, respectively (fig. S4). (B) Side-chain
mobility of the C[4]-symmetric Rua, color-coded from 0°
(blue) to 90° (red) angular spread in the torsion angles plots at various
scales with mutated residues as colored spheres. (A) Sketch of
an asymmetric interface between patches a and b, which, in
general, gives rise to an infinite helix (top). A C[2]-symmetric
interface also between patches a and b doubles the numbers of
contacts and forms a globular complex (bottom). Along the same
lines, the reported D[2], D[4], and D[8] oligomers have 4-, 8-,
and 16-fold contacts, respectively (fig. S4). (B) Side-chain
mobility of the C[4]-symmetric Rua, color-coded from 0°
(blue) to 90° (red) angular spread in the torsion angles
 [1] and [1] and  [2]
(24). The C- and N-terminal domains are at the top and bottom,
respectively. (C) Pga-A and -B designed in crystal contact
a-a(25). (D) Pga-C and-D designed in crystal contact f-f (25).
(E) Oas-A and-B planned as a D[2] tetramer at a rotation angle
of 86° around a common molecular twofold axis (vertical).
(F) Oas-C designed as a D[2] tetramer at an alternative rotation
angle of 29°. (G) Designed D[2] tetramer of Uro-A around a
common molecular twofold axis (vertical). The designed contact
is between the NAD^+-binding domains (residues 142 to 343),
which are given in lighter hues. (H) Designed octameric Rua-D
with a head-head contact. [2]
(24). The C- and N-terminal domains are at the top and bottom,
respectively. (C) Pga-A and -B designed in crystal contact
a-a(25). (D) Pga-C and-D designed in crystal contact f-f (25).
(E) Oas-A and-B planned as a D[2] tetramer at a rotation angle
of 86° around a common molecular twofold axis (vertical).
(F) Oas-C designed as a D[2] tetramer at an alternative rotation
angle of 29°. (G) Designed D[2] tetramer of Uro-A around a
common molecular twofold axis (vertical). The designed contact
is between the NAD^+-binding domains (residues 142 to 343),
which are given in lighter hues. (H) Designed octameric Rua-D
with a head-head contact.
|
 |
Figure 3.
Fig. 3. Established oligomer structures (24). All mutations are
marked by purple spheres. (A) Crystal structure of
C[2]-symmetric Uro-A showing the twofold molecular symmetry axis
(red) and four local twofold axes relating the cores (darker
colors) and the NAD^+ domains (light colors) to their
counterparts. The interface between core and NAD^+ domains was
broken in the lower left and upper right chains. (B)
D[4]-symmetric octamer Rua-A. (C) C[2]-symmetric octamer Rua-B.
(D) Negatively stained electron micrograph of Rua-E showing the
fiber association and a Rua-A octamer (B) at the scale defined
by the box edge. (E) Native mycobacterial porin (28). The
encircled membrane-immersed part was deleted, giving rise to
Myp-A. (F) D[8]-symmetric association of two Myp-A molecules
(top and bottom ring). The positions of the 52-residue deletions
are marked by red spheres (fig. S1D).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2008,
319,
206-209)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.E.Schulz
(2011).
A new classification of membrane protein crystals.
|
| |
J Mol Biol,
407,
640-646.
|
 |
|
|
|
|
 |
G.J.Forse,
N.Ram,
D.R.Banatao,
D.Cascio,
M.R.Sawaya,
H.E.Klock,
S.A.Lesley,
and
T.O.Yeates
(2011).
Synthetic symmetrization in the crystallization and structure determination of CelA from Thermotoga maritima.
|
| |
Protein Sci,
20,
168-178.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Onoda,
Y.Ueya,
T.Sakamoto,
T.Uematsu,
and
T.Hayashi
(2010).
Supramolecular hemoprotein-gold nanoparticle conjugates.
|
| |
Chem Commun (Camb),
46,
9107-9109.
|
 |
|
|
|
|
 |
C.Fan,
S.Cheng,
Y.Liu,
C.M.Escobar,
C.S.Crowley,
R.E.Jefferson,
T.O.Yeates,
and
T.A.Bobik
(2010).
Short N-terminal sequences package proteins into bacterial microcompartments.
|
| |
Proc Natl Acad Sci U S A,
107,
7509-7514.
|
 |
|
|
|
|
 |
K.Hashimoto,
and
A.R.Panchenko
(2010).
Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states.
|
| |
Proc Natl Acad Sci U S A,
107,
20352-20357.
|
 |
|
|
|
|
 |
M.Biancalana,
K.Makabe,
and
S.Koide
(2010).
Minimalist design of water-soluble cross-beta architecture.
|
| |
Proc Natl Acad Sci U S A,
107,
3469-3474.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Mitchell,
A.Ebner,
P.Hinterdorfer,
R.Tampé,
and
S.Howorka
(2010).
Chemical tags mediate the orthogonal self-assembly of DNA duplexes into supramolecular structures.
|
| |
Small,
6,
1732-1735.
|
 |
|
|
|
|
 |
N.Yokoi,
H.Inaba,
M.Terauchi,
A.Z.Stieg,
N.J.Sanghamitra,
T.Koshiyama,
K.Yutani,
S.Kanamaru,
F.Arisaka,
T.Hikage,
A.Suzuki,
T.Yamane,
J.K.Gimzewski,
Y.Watanabe,
S.Kitagawa,
and
T.Ueno
(2010).
Construction of robust bio-nanotubes using the controlled self-assembly of component proteins of bacteriophage T4.
|
| |
Small,
6,
1873-1879.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Papapostolou,
and
S.Howorka
(2009).
Engineering and exploiting protein assemblies in synthetic biology.
|
| |
Mol Biosyst,
5,
723-732.
|
 |
|
|
|
|
 |
J.Wang,
T.Palzkill,
and
D.C.Chow
(2009).
Structural insight into the kinetics and DeltaCp of interactions between TEM-1 beta-lactamase and beta-lactamase inhibitory protein (BLIP).
|
| |
J Biol Chem,
284,
595-609.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bhattacharyya,
and
S.Vishveshwara
(2009).
Functional correlation of bacterial LuxS with their quaternary associations: interface analysis of the structure networks.
|
| |
BMC Struct Biol,
9,
8.
|
 |
|
|
|
|
 |
M.Montal
(2009).
Vpu matchmakers as a therapeutic strategy for HIV infection.
|
| |
PLoS Pathog,
5,
e1000246.
|
 |
|
|
|
|
 |
N.Jouravel,
E.Sablin,
M.Togashi,
J.D.Baxter,
P.Webb,
and
R.J.Fletterick
(2009).
Molecular basis for dimer formation of TRbeta variant D355R.
|
| |
Proteins,
75,
111-117.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.B.Crowley,
P.M.Matias,
A.R.Khan,
M.Roessle,
and
D.I.Svergun
(2009).
Metal-mediated self-assembly of a beta-sandwich protein.
|
| |
Chemistry,
15,
12672-12680.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.A.Whitehead,
E.Je,
and
D.S.Clark
(2009).
Rational shape engineering of the filamentous protein gamma prefoldin through incremental gene truncation.
|
| |
Biopolymers,
91,
496-503.
|
 |
|
|
|
|
 |
Y.Wine,
N.Cohen-Hadar,
R.Lamed,
A.Freeman,
and
F.Frolow
(2009).
Modification of protein crystal packing by systematic mutations of surface residues: implications on biotemplating and crystal porosity.
|
| |
Biotechnol Bioeng,
104,
444-457.
|
 |
|
|
|
|
 |
D.Marsic,
R.C.Hughes,
M.L.Byrne-Steele,
and
J.D.Ng
(2008).
PCR-based gene synthesis to produce recombinant proteins for crystallization.
|
| |
BMC Biotechnol,
8,
44.
|
 |
|
|
|
|
 |
E.D.Levy,
E.Boeri Erba,
C.V.Robinson,
and
S.A.Teichmann
(2008).
Assembly reflects evolution of protein complexes.
|
| |
Nature,
453,
1262-1265.
|
 |
|
|
|
|
 |
E.N.Salgado,
R.A.Lewis,
J.Faraone-Mennella,
and
F.A.Tezcan
(2008).
Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
|
| |
J Am Chem Soc,
130,
6082-6084.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.F.Chou,
C.So,
B.R.White,
J.C.Carlson,
M.Sarikaya,
and
C.R.Wagner
(2008).
Enzyme nanorings.
|
| |
ACS Nano,
2,
2519-2525.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

