 |
PDBsum entry 2std

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.94
- scytalone dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
scytalone = 1,3,8-trihydroxynaphthalene + H2O
|
 |
 |
 |
 |
 |
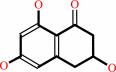 scytalone
scytalone
|
=
|
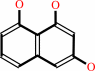 1,3,8-trihydroxynaphthalene
1,3,8-trihydroxynaphthalene
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:9931-9939
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cryogenic X-ray crystal structure analysis for the complex of scytalone dehydratase of a rice blast fungus and its tight-binding inhibitor, carpropamid: the structural basis of tight-binding inhibition.
|
|
M.Nakasako,
T.Motoyama,
Y.Kurahashi,
I.Yamaguchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Scytalone dehydratase is a member of the group of enzymes involved in fungal
melanin biosynthesis in a phytopathogenic fungus, Pyricularia oryzae, which
causes rice blast disease. Carpropamid [(1RS,3SR)-2,
2-dichloro-N-[(R)-1-(4-chlorophenyl)ethyl]-1-ethyl-3-methylcyclopropa
necarboxamide] is a tight-binding inhibitor of the enzyme. To clarify the
structural basis for tight-binding inhibition, the crystal structure of the
enzyme complexed with carpropamid was analyzed using diffraction data collected
at 100 K. The structural model was refined to a crystallographic R-factor of
0.180 against reflections up to a resolution of 2.1 A. Carpropamid was bound in
a hydrophobic cavity of the enzyme. Three types of interactions appeared to
contribute to the binding. (i) A hydrogen bond was formed between a chloride
atom in the dichloromethylethylcyclopropane ring of carpropamid and Asn-131 of
the enzyme. (ii) The (chlorophenyl)ethyl group of carpropamid built strong
contacts with Val-75, and this group further formed a cluster of aromatic rings
together with four aromatic residues in the enzyme (Tyr-50, Phe-53, Phe-158, and
Phe-162). (iii) Two hydration water molecules bound to the carboxamide group of
carpropamid, and they were further hydrogen-bonded to Tyr-30, Tyr-50, His-85,
and His-110. As a result of interactions between carpropamid and the
phenylalanine residues (Phe-158 and Phe-162) in the C-terminal region of the
enzyme, the C-terminal region completely covered the inhibitor, ensuring its
localization in the cavity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
V.Bhadauria,
L.X.Wang,
and
Y.L.Peng
(2010).
Proteomic changes associated with deletion of the Magnaporthe oryzae conidial morphology-regulating gene COM1.
|
| |
Biol Direct,
5,
61.
|
 |
|
|
|
|
 |
D.S.Kim,
Y.M.Jeong,
I.K.Park,
H.G.Hahn,
H.K.Lee,
S.B.Kwon,
J.H.Jeong,
S.J.Yang,
U.D.Sohn,
and
K.C.Park
(2007).
A new 2-imino-1,3-thiazoline derivative, KHG22394, inhibits melanin synthesis in mouse B16 melanoma cells.
|
| |
Biol Pharm Bull,
30,
180-183.
|
 |
|
|
|
|
 |
M.Nakasako
(2004).
Water-protein interactions from high-resolution protein crystallography.
|
| |
Philos Trans R Soc Lond B Biol Sci,
359,
1191.
|
 |
|
|
|
|
 |
N.Yamada,
T.Motoyama,
M.Nakasako,
S.Kagabu,
T.Kudo,
and
I.Yamaguchi
(2004).
Enzymatic characterization of scytalone dehydratase Val75Met variant found in melanin biosynthesis dehydratase inhibitor (MBI-D) resistant strains of the rice blast fungus.
|
| |
Biosci Biotechnol Biochem,
68,
615-621.
|
 |
|
|
|
|
 |
D.B.Jordan,
and
G.S.Basarab
(2000).
Binding dynamics of two water molecules constrained within the scytalone dehydratase binding pocket.
|
| |
Bioorg Med Chem Lett,
10,
23-26.
|
 |
|
|
|
|
 |
A.E.Nixon,
S.M.Firestine,
F.G.Salinas,
and
S.J.Benkovic
(1999).
Rational design of a scytalone dehydratase-like enzyme using a structurally homologous protein scaffold.
|
| |
Proc Natl Acad Sci U S A,
96,
3568-3571.
|
 |
|
|
|
|
 |
G.S.Basarab,
D.B.Jordan,
T.C.Gehret,
R.S.Schwartz,
and
Z.Wawrzak
(1999).
Design of scytalone dehydratase inhibitors as rice blast fungicides: derivatives of norephedrine.
|
| |
Bioorg Med Chem Lett,
9,
1613-1618.
|
 |
|
|
|
|
 |
Z.Wawrzak,
T.Sandalova,
J.J.Steffens,
G.S.Basarab,
T.Lundqvist,
Y.Lindqvist,
and
D.B.Jordan
(1999).
High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
|
| |
Proteins,
35,
425-439.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

