 |
PDBsum entry 2j1e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.169
- protein O-GlcNAcase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
3-O-(N-acetyl-beta-D-glucosaminyl)-L-seryl-[protein] + H2O = N-acetyl-D-glucosamine + L-seryl-[protein]
|
|
2.
|
3-O-(N-acetyl-beta-D-glucosaminyl)-L-threonyl-[protein] + H2O = L-threonyl-[protein] + N-acetyl-D-glucosamine
|
|
 |
 |
 |
 |
 |
3-O-(N-acetyl-beta-D-glucosaminyl)-L-seryl-[protein]
|
+
|
H2O
|
=
|
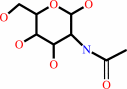 N-acetyl-D-glucosamine
N-acetyl-D-glucosamine
|
+
|
L-seryl-[protein]
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
3-O-(N-acetyl-beta-D-glucosaminyl)-L-threonyl-[protein]
|
+
|
H2O
|
=
|
L-threonyl-[protein]
|
+
|
N-acetyl-D-glucosamine
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:37748-37757
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The interaction of a carbohydrate-binding module from a Clostridium perfringens N-acetyl-beta-hexosaminidase with its carbohydrate receptor.
|
|
E.Ficko-Blean,
A.B.Boraston.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Clostridium perfringens is a notable colonizer of the human gastrointestinal
tract. This bacterium is quite remarkable for a human pathogen by the number of
glycoside hydrolases found in its genome. The modularity of these enzymes is
striking as is the frequent occurrence of modules having amino acid sequence
identity with family 32 carbohydrate-binding modules (CBMs), often referred to
as F5/8 domains. Here we report the properties of family 32 CBMs from a C.
perfringens N-acetyl-beta-hexosaminidase. Macroarray, UV difference, and
isothermal titration calorimetry binding studies indicate a preference for the
disaccharide LacNAc (beta-d-galactosyl-1,4-beta-d-N-acetylglucosamine). The
molecular details of the interaction of this CBM with galactose, LacNAc, and the
type II blood group H-trisaccharide are revealed by x-ray crystallographic
studies at resolutions of 1.49, 2.4, and 2.3 A, respectively.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIGURE 4. Representative electron density and interaction
of CpCBM32 with galactose (A), LacNAc (B), and the type II blood
group H-trisaccharide (C). All maps are maximum-likelihood/  [A]
(37)-weighted 2F[obs] - F[calc] electron density maps contoured
at 1 [A]
(37)-weighted 2F[obs] - F[calc] electron density maps contoured
at 1  (0.35, 0.30, and 0.29
electrons/Å^3 for galactose, LacNAc, and the
H-trisaccharide, respectively). The disordered loop in the
galactose complex is shown with a dashed line. Amino acid side
chains involved in binding the sugars are shown in gray stick
representation and labeled. (0.35, 0.30, and 0.29
electrons/Å^3 for galactose, LacNAc, and the
H-trisaccharide, respectively). The disordered loop in the
galactose complex is shown with a dashed line. Amino acid side
chains involved in binding the sugars are shown in gray stick
representation and labeled.
|
 |
Figure 5.
FIGURE 5. Schematics showing the interactions of CpCBM32
with galactose (A), LacNAc (B), and the type II blood group
H-trisaccharide (C). A distance of 3.2 Å was used as the
cut-off for determination of significant hydrogen bonds. Water
molecules are shown as shaded spheres.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
37748-37757)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Guillén,
S.Sánchez,
and
R.Rodríguez-Sanoja
(2010).
Carbohydrate-binding domains: multiplicity of biological roles.
|
| |
Appl Microbiol Biotechnol,
85,
1241-1249.
|
 |
|
|
|
|
 |
D.W.Abbott,
M.A.Higgins,
S.Hyrnuik,
B.Pluvinage,
A.Lammerts van Bueren,
and
A.B.Boraston
(2010).
The molecular basis of glycogen breakdown and transport in Streptococcus pneumoniae.
|
| |
Mol Microbiol,
77,
183-199.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Fushinobu
(2010).
Unique sugar metabolic pathways of bifidobacteria.
|
| |
Biosci Biotechnol Biochem,
74,
2374-2384.
|
 |
|
|
|
|
 |
A.Peer,
S.P.Smith,
E.A.Bayer,
R.Lamed,
and
I.Borovok
(2009).
Noncellulosomal cohesin- and dockerin-like modules in the three domains of life.
|
| |
FEMS Microbiol Lett,
291,
1.
|
 |
|
|
|
|
 |
E.Ficko-Blean,
and
A.B.Boraston
(2009).
N-acetylglucosamine recognition by a family 32 carbohydrate-binding module from Clostridium perfringens NagH.
|
| |
J Mol Biol,
390,
208-220.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Ficko-Blean,
K.J.Gregg,
J.J.Adams,
J.H.Hehemann,
M.Czjzek,
S.P.Smith,
and
A.B.Boraston
(2009).
Portrait of an enzyme, a complete structural analysis of a multimodular {beta}-N-acetylglucosaminidase from Clostridium perfringens.
|
| |
J Biol Chem,
284,
9876-9884.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Koropatkin,
E.C.Martens,
J.I.Gordon,
and
T.J.Smith
(2009).
Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont.
|
| |
Biochemistry,
48,
1532-1542.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.M.Cheng,
F.C.Hsieh,
and
M.Meng
(2009).
Functional analysis of conserved aromatic amino acids in the discoidin domain of Paenibacillus beta-1,3-glucanase.
|
| |
Microb Cell Fact,
8,
62.
|
 |
|
|
|
|
 |
E.Ficko-Blean,
K.A.Stubbs,
O.Nemirovsky,
D.J.Vocadlo,
and
A.B.Boraston
(2008).
Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB.
|
| |
Proc Natl Acad Sci U S A,
105,
6560-6565.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.J.Stevens
(2008).
Possible evolutionary links between immunoglobulin light chains and other proteins involved in amyloidosis.
|
| |
Amyloid,
15,
96.
|
 |
|
|
|
|
 |
H.Brüggemann,
and
G.Gottschalk
(2008).
Comparative genomics of clostridia: link between the ecological niche and cell surface properties.
|
| |
Ann N Y Acad Sci,
1125,
73-81.
|
 |
|
|
|
|
 |
J.J.Adams,
K.Gregg,
E.A.Bayer,
A.B.Boraston,
and
S.P.Smith
(2008).
Structural basis of Clostridium perfringens toxin complex formation.
|
| |
Proc Natl Acad Sci U S A,
105,
12194-12199.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Wada,
T.Ando,
M.Kiyohara,
H.Ashida,
M.Kitaoka,
M.Yamaguchi,
H.Kumagai,
T.Katayama,
and
K.Yamamoto
(2008).
Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure.
|
| |
Appl Environ Microbiol,
74,
3996-4004.
|
 |
|
|
|
|
 |
K.J.Gregg,
R.Finn,
D.W.Abbott,
and
A.B.Boraston
(2008).
Divergent modes of glycan recognition by a new family of carbohydrate-binding modules.
|
| |
J Biol Chem,
283,
12604-12613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Nakajima,
T.Nihira,
M.Nishimoto,
and
M.Kitaoka
(2008).
Identification of galacto-N-biose phosphorylase from Clostridium perfringens ATCC13124.
|
| |
Appl Microbiol Biotechnol,
78,
465-471.
|
 |
|
|
|
|
 |
O.Okhrimenko,
and
I.Jelesarov
(2008).
A survey of the year 2006 literature on applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
21,
1.
|
 |
|
|
|
|
 |
R.M.Weiner,
L.E.Taylor,
B.Henrissat,
L.Hauser,
M.Land,
P.M.Coutinho,
C.Rancurel,
E.H.Saunders,
A.G.Longmire,
H.Zhang,
E.A.Bayer,
H.J.Gilbert,
F.Larimer,
I.B.Zhulin,
N.A.Ekborg,
R.Lamed,
P.M.Richardson,
I.Borovok,
and
S.Hutcheson
(2008).
Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T.
|
| |
PLoS Genet,
4,
e1000087.
|
 |
|
|
|
|
 |
J.Finsterer,
and
B.Hess
(2007).
Neuromuscular and central nervous system manifestations of Clostridium perfringens infections.
|
| |
Infection,
35,
396-405.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

