 |
PDBsum entry 2hgx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of cys315ala mutant of human mitochondrial branched chain aminotransferase
|
|
Structure:
|
 |
Branched-chain-amino-acid aminotransferase, mitochondrial. Chain: a, b. Synonym: bcatm, placental protein 18, pp18, mitochondrial branched chain aminotransferase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: bcat2, bcatm, eca40. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.208
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
N.H.Yennawar,S.M.Hutson
|
Key ref:

|
 |
N.H.Yennawar
et al.
(2006).
Human mitochondrial branched chain aminotransferase isozyme: structural role of the CXXC center in catalysis.
J Biol Chem,
281,
39660-39671.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
27-Jun-06
|
Release date:
|
24-Oct-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O15382
(BCAT2_HUMAN) -
Branched-chain-amino-acid aminotransferase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
392 a.a.
357 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.42
- branched-chain-amino-acid transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-leucine + 2-oxoglutarate = 4-methyl-2-oxopentanoate + L-glutamate
|
 |
 |
 |
 |
 |
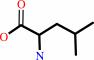 L-leucine
L-leucine
|
+
|
2-oxoglutarate
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
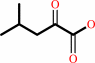 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:39660-39671
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Human mitochondrial branched chain aminotransferase isozyme: structural role of the CXXC center in catalysis.
|
|
N.H.Yennawar,
M.M.Islam,
M.Conway,
R.Wallin,
S.M.Hutson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mammalian branched chain aminotransferases (BCATs) have a unique CXXC center.
Kinetic and structural studies of three CXXC center mutants (C315A, C318A, and
C315A/C318A) of human mitochondrial (hBCATm) isozyme and the oxidized hBCATm
enzyme (hBCATm-Ox) have been used to elucidate the role of this center in hBCATm
catalysis. X-ray crystallography revealed that the CXXC motif, through its
network of hydrogen bonds, plays a crucial role in orienting the substrate
optimally for catalysis. In all structures, there were changes in the structure
of the beta-turn preceding the CXXC motif when compared with wild type protein.
The N-terminal loop between residues 15 and 32 is flexible in the oxidized and
mutant enzymes, the disorder greater in the oxidized protein. Disordering of the
N-terminal loop disrupts the integrity of the side chain binding pocket,
particularly for the branched chain side chain, less so for the dicarboxylate
substrate side chain. The kinetic studies of the mutant and oxidized enzymes
support the structural analysis. The kinetic results showed that the predominant
effect of oxidation was on the second half-reaction rather than the first
half-reaction. The oxidized enzyme was completely inactive, whereas the mutants
showed limited activity. Model building of the second half-reaction substrate
alpha-ketoisocaproate in the pyridoxamine 5'-phosphate-hBCATm structure suggests
that disruption of the CXXC center results in altered substrate orientation and
deprotonation of the amino group of pyridoxamine 5'-phosphate, which inhibits
catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. C XX C center of oxidized and mutant hBCATm
proteins. A, oxidized WT-hBCATm; B, C315A hBCATm; C, C315A/C318A
double mutant hBCATm. In the reduced WT protein, the sulfurs of
Cys^315 and Cys^318 form a thiol-thiolate hydrogen bond. Upon
mutation or oxidation, the thiol-thiolate interaction does not
exist. Instead, the Cys^315 and Cys^318 sulfurs form a disulfide
bridge.
|
 |
Figure 4.
FIGURE 4. Active site structures of C315A mutant hBCATm in
complex with N-methylleucine (C315A-NML). A and B show the 2F[o]
- F[c] difference Fourier electron density maps of the active
site in monomer A and monomer B, respectively. C, the
superimposed model of the N-methylleucine-bound WT-hBCATm
structure and structure of N-methylleucine-bound C315A mutant
hBCATm. The carbon skeletons are in khaki in WT-hBCATm and in
green in the single mutant C315A-NML structures.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
39660-39671)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Brunetti-Pierri,
B.Lanpher,
A.Erez,
E.A.Ananieva,
M.Islam,
J.C.Marini,
Q.Sun,
C.Yu,
M.Hegde,
J.Li,
R.M.Wynn,
D.T.Chuang,
S.Hutson,
and
B.Lee
(2011).
Phenylbutyrate therapy for maple syrup urine disease.
|
| |
Hum Mol Genet,
20,
631-640.
|
 |
|
|
|
|
 |
A.Castell,
C.Mille,
and
T.Unge
(2010).
Structural analysis of mycobacterial branched-chain aminotransferase: implications for inhibitor design.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
549-557.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.M.Islam,
M.Nautiyal,
R.M.Wynn,
J.A.Mobley,
D.T.Chuang,
and
S.M.Hutson
(2010).
Branched-chain amino acid metabolon: interaction of glutamate dehydrogenase with the mitochondrial branched-chain aminotransferase (BCATm).
|
| |
J Biol Chem,
285,
265-276.
|
 |
|
|
|
|
 |
M.Nautiyal,
A.J.Sweatt,
J.A.MacKenzie,
R.Mark Payne,
S.Szucs,
R.Matalon,
R.Wallin,
and
S.M.Hutson
(2010).
Neuronal localization of the mitochondrial protein NIPSNAP1 in rat nervous system.
|
| |
Eur J Neurosci,
32,
560-569.
|
 |
|
|
|
|
 |
L.W.Tremblay,
and
J.S.Blanchard
(2009).
The 1.9 A structure of the branched-chain amino-acid transaminase (IlvE) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1071-1077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Murín,
and
B.Hamprecht
(2008).
Metabolic and regulatory roles of leucine in neural cells.
|
| |
Neurochem Res,
33,
279-284.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

