 |
PDBsum entry 2gcu

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
X-ray structure of gene product from arabidopsis thaliana at1g53580
|
|
Structure:
|
 |
Putative hydroxyacylglutathione hydrolase 3. Chain: a, b, c, d. Synonym: glyoxalase ii, glx ii. Engineered: yes
|
|
Source:
|
 |
Arabidopsis thaliana. Thale cress. Organism_taxid: 3702. Gene: at1g53580. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
1.48Å
|
R-factor:
|
0.178
|
R-free:
|
0.204
|
|
|
Authors:
|
 |
J.G.Mccoy,G.E.Wesenberg,G.N.Phillips Jr.,E.Bitto,C.A.Bingman,Center For Eukaryotic Structural Genomics (Cesg)
|
Key ref:

|
 |
J.G.McCoy
et al.
(2006).
Structure of an ETHE1-like protein from Arabidopsis thaliana.
Acta Crystallogr D Biol Crystallogr,
62,
964-970.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-Mar-06
|
Release date:
|
18-Apr-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9C8L4
(ETHE1_ARATH) -
Persulfide dioxygenase ETHE1 homolog, mitochondrial from Arabidopsis thaliana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
294 a.a.
244 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.18
- persulfide dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-sulfanylglutathione + O2 + H2O = sulfite + glutathione + 2 H+
|
 |
 |
 |
 |
 |
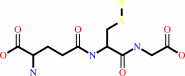 S-sulfanylglutathione
S-sulfanylglutathione
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
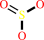 sulfite
sulfite
|
+
|
glutathione
Bound ligand (Het Group name = )
matches with 80.00% similarity
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:964-970
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of an ETHE1-like protein from Arabidopsis thaliana.
|
|
J.G.McCoy,
C.A.Bingman,
E.Bitto,
M.M.Holdorf,
C.A.Makaroff,
G.N.Phillips.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The protein product of gene At1g53580 from Arabidopsis thaliana possesses 54%
sequence identity to a human enzyme that has been implicated in the rare
disorder ethylmalonic encephalopathy. The structure of the At1g53580 protein has
been solved to a nominal resolution of 1.48 Angstrom. This structure reveals
tertiary structure differences between the ETHE1-like enzyme and glyoxalase II
enzymes that are likely to account for differences in reaction chemistry and
multimeric state between the two types of enzymes. In addition, the Arabidopsis
ETHE1 protein is used as a model to explain the significance of several
mutations in the human enzyme that have been observed in patients with
ethylmalonic encephalopathy.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 (a) The At1g53580 monomer. Helices are labelled 1-8
and  -strands
are labelled A-L. Residues known to be involved in human
ethylmalonic encephalopathy in human ETHE1 are colored pink. (b)
Overlay of the AtETHE1 (cyan) and the GLX2-5 (magenta) monomers.
The metal ions from the GLX2-5 structure are colored orange and
the iron ion from the AtETHE1 structure is colored gray. Arrows
point to changes between the folds of the two enzymes. -strands
are labelled A-L. Residues known to be involved in human
ethylmalonic encephalopathy in human ETHE1 are colored pink. (b)
Overlay of the AtETHE1 (cyan) and the GLX2-5 (magenta) monomers.
The metal ions from the GLX2-5 structure are colored orange and
the iron ion from the AtETHE1 structure is colored gray. Arrows
point to changes between the folds of the two enzymes.
|
 |
Figure 5.
Figure 5 Overlay of the substrate-binding residues from the
human glyoxalase II (cyan) with the equivalent residues in the
AtETHE1 enzyme (magenta). S-Hydroxybromophenylcarbamoyl
glutathione bound to glyoxalase II is colored yellow. Labels
correspond to AtETHE1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
964-970)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.L.Stamp,
P.Owen,
K.E.Omari,
C.E.Nichols,
M.Lockyer,
H.K.Lamb,
I.G.Charles,
A.R.Hawkins,
and
D.K.Stammers
(2010).
Structural and functional characterization of Salmonella enterica serovar Typhimurium YcbL: an unusual Type II glyoxalase.
|
| |
Protein Sci,
19,
1897-1905.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Limphong,
G.Nimako,
P.W.Thomas,
W.Fast,
C.A.Makaroff,
and
M.W.Crowder
(2009).
Arabidopsis thaliana mitochondrial glyoxalase 2-1 exhibits beta-lactamase activity.
|
| |
Biochemistry,
48,
8491-8493.
|
 |
|
|
|
|
 |
P.Limphong,
M.W.Crowder,
B.Bennett,
and
C.A.Makaroff
(2009).
Arabidopsis thaliana GLX2-1 contains a dinuclear metal binding site, but is not a glyoxalase 2.
|
| |
Biochem J,
417,
323-330.
|
 |
|
|
|
|
 |
V.Tiranti,
C.Viscomi,
T.Hildebrandt,
I.Di Meo,
R.Mineri,
C.Tiveron,
M.D.Levitt,
A.Prelle,
G.Fagiolari,
M.Rimoldi,
and
M.Zeviani
(2009).
Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy.
|
| |
Nat Med,
15,
200-205.
|
 |
|
|
|
|
 |
M.M.Holdorf,
B.Bennett,
M.W.Crowder,
and
C.A.Makaroff
(2008).
Spectroscopic studies on Arabidopsis ETHE1, a glyoxalase II-like protein.
|
| |
J Inorg Biochem,
102,
1825-1830.
|
 |
|
|
|
|
 |
G.N.Phillips,
B.G.Fox,
J.L.Markley,
B.F.Volkman,
E.Bae,
E.Bitto,
C.A.Bingman,
R.O.Frederick,
J.G.McCoy,
B.L.Lytle,
B.S.Pierce,
J.Song,
and
S.N.Twigger
(2007).
Structures of proteins of biomedical interest from the Center for Eukaryotic Structural Genomics.
|
| |
J Struct Funct Genomics,
8,
73-84.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

