 |
PDBsum entry 2dkc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of n-acetylglucosamine-phosphate mutase, a member of the alpha-d-phosphohexomutase superfamily, in the substrate complex
|
|
Structure:
|
 |
Phosphoacetylglucosamine mutase. Chain: a, b. Synonym: pagm, acetylglucosamine phosphomutase, n-acetylglucosamine- phosphate mutase. Engineered: yes
|
|
Source:
|
 |
Candida albicans. Organism_taxid: 5476. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.189
|
R-free:
|
0.240
|
|
|
Authors:
|
 |
Y.Nishitani,D.Maruyama,T.Nonaka,A.Kita,T.A.Fukami,T.Mio,H.Yamada- Okabe,T.Yamada-Okabe,K.Miki
|
Key ref:

|
 |
Y.Nishitani
et al.
(2006).
Crystal structures of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, and its substrate and product complexes.
J Biol Chem,
281,
19740-19747.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Apr-06
|
Release date:
|
16-May-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9P4V2
(AGM1_CANAX) -
Phosphoacetylglucosamine mutase from Candida albicans
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
544 a.a.
536 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.2.3
- phosphoacetylglucosamine mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
UDP-N-acetylglucosamine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-acetyl-alpha-D-glucosamine 1-phosphate = N-acetyl-D-glucosamine 6-phosphate
|
 |
 |
 |
 |
 |
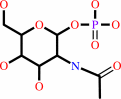 N-acetyl-alpha-D-glucosamine 1-phosphate
N-acetyl-alpha-D-glucosamine 1-phosphate
|
=
|
N-acetyl-D-glucosamine 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:19740-19747
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, and its substrate and product complexes.
|
|
Y.Nishitani,
D.Maruyama,
T.Nonaka,
A.Kita,
T.A.Fukami,
T.Mio,
H.Yamada-Okabe,
T.Yamada-Okabe,
K.Miki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
N-acetylglucosamine-phosphate mutase (AGM1) is an essential enzyme in the
synthetic process of UDP-N-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is a UDP
sugar that serves as a biosynthetic precursor of glycoproteins,
mucopolysaccharides, and the cell wall of bacteria. Thus, a specific inhibitor
of AGM1 from pathogenetic fungi could be a new candidate for an antifungal
reagent that inhibits cell wall synthesis. AGM1 catalyzes the conversion of
N-acetylglucosamine 6-phosphate (GlcNAc-6-P) into N-acetylglucosamine
1-phosphate (GlcNAc-1-P). This enzyme is a member of the
alpha-D-phosphohexomutase superfamily, which catalyzes the intramolecular
phosphoryl transfer of sugar substrates. Here we report the crystal structures
of AGM1 from Candida albicans for the first time, both in the apoform and in the
complex forms with the substrate and the product, and discuss its catalytic
mechanism. The structure of AGM1 consists of four domains, of which three
domains have essentially the same fold. The overall structure is similar to
those of phosphohexomutases; however, there are two additional beta-strands in
domain 4, and a circular permutation occurs in domain 1. The catalytic cleft is
formed by four loops from each domain. The N-acetyl group of the substrate is
recognized by Val-370 and Asn-389 in domain 3, from which the substrate
specificity arises. By comparing the substrate and product complexes, it is
suggested that the substrate rotates about 180 degrees on the axis linking C-4
and the midpoint of the C-5-O-5 bond in the reaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Schematic illustration of the role of AGM1 in the
biosynthetic pathway of UDP-GlcNAc. In eukaryotes, UDP-GlcNAc is
synthesized from fructose 6-phosphate by four successive
reactions: (i) the conversion of Fru-6-P into GlcN-6-P; (ii) the
acetylation of GlcN-6-P into GlcNAc-6-P; (iii) the
interconversion of GlcNAc-6-P and GlcNAc-1-P; and (iv) the
uridylation of GlcNAc-1-P into UDP-GlcNAc. AGM1 is catalyzed in
step iii.In prokaryotes, the intramolecular phosphoryl transfer
(step iii) occurs before acetylation (step ii), and GlcN-1-P is
generated as the intermediate.
|
 |
Figure 8.
Schematic drawing of the proposed catalytic mechanism for the
conversion of GlcNAc-6-P to GlcNAc-1-P by AGM1. Phosphoryl
groups are indicated by P in a shaded circle. The phosphoryl
group near Ser-66 first binds to the substrate. The substrate is
converted into a bis-phosphorylated intermediate. Then, this
intermediate rotates at 180°. Consequently, the phosphoryl
group at C-6 changes positions with another phosphoryl group at
C-1. Finally, the phosphoryl group moved near the metal ion
dissociates from the intermediate and binds to Ser-66.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
19740-19747)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Resch,
E.Schiltz,
F.Titgemeyer,
and
Y.A.Muller
(2010).
Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA.
|
| |
Nucleic Acids Res,
38,
2485-2497.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Barreteau,
A.Kovac,
A.Boniface,
M.Sova,
S.Gobec,
and
D.Blanot
(2008).
Cytoplasmic steps of peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
168-207.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

