 |
PDBsum entry 2ctb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
HydrolasE(C-terminal peptidase)
|
PDB id
|
|

|

|
2ctb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.17.1
- carboxypeptidase A.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Peptidyl-L-amino acid + H2O = peptide + L-amino acid
|
 |
 |
 |
 |
 |
|
+
|
|
=
|
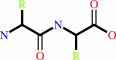
|
+
|
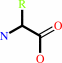
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
49:534-540
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
High-resolution structure of the complex between carboxypeptidase A and L-phenyl lactate.
|
|
A.Teplyakov,
K.S.Wilson,
P.Orioli,
S.Mangani.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray structures of native carboxypeptidase A and of the enzyme-inhibitor
complex with L-phenyl lactate have been refined at 1.54 and 1.45 A resolution to
R factors of 0.151 and 0.161, respectively. Crystals of the complex were
isomorphous with the native crystals (space group P2(1), a = 51.60, b = 60.27, c
= 47.25 A, beta = 97.27 degrees ). The high-resolution electron density allowed
correction of many side-chain positions in the classical carboxypeptidase A
model. This reflects the advantages of the high-quality complete synchrotron
data collected with an imaging plate detector. The conformational changes in the
active centre of the enzyme upon binding of the inhibitor are restricted to only
two residues, Tyr248 and Arg145. L-Phenyl lactate is bound in the S1' pocket and
forms hydrogen bonds to Arg145, Glu270 and to the zinc-bound water molecule. The
present structure provides an explanation for the higher stability of the
complexes with the products of esterolysis in comparison with those of
amidolysis. This is consistent with the finding that product release is rate
limiting for esters but not for peptides.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. The ~o/~ plot for the native CPA model.
|
 |
Figure 3.
Fig. 3. Representaive fragment of the (3Fo 2Fc) lectron density of CPA contoured at the level of 1.Str, where o is the
rootmeansquare deviation. The present CPA model is shown in green, the 5CPA odel is shown in orange. (a) Ser254 and Ile255; (b)
Leu271 and Tyr238 with the hydrogenbonded water molecule.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(1993,
49,
534-540)
copyright 1993.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Koike,
A.Kidera,
and
M.Ota
(2009).
Alteration of oligomeric state and domain architecture is essential for functional transformation between transferase and hydrolase with the same scaffold.
|
| |
Protein Sci,
18,
2060-2066.
|
 |
|
|
|
|
 |
M.A.Dolan,
M.Keil,
and
D.S.Baker
(2008).
Comparison of composer and ORCHESTRAR.
|
| |
Proteins,
72,
1243-1258.
|
 |
|
|
|
|
 |
K.H.Kim
(2007).
Outliers in SAR and QSAR: is unusual binding mode a possible source of outliers?
|
| |
J Comput Aided Mol Des,
21,
63-86.
|
 |
|
|
|
|
 |
W.Cai,
J.Pei,
and
N.V.Grishin
(2004).
Reconstruction of ancestral protein sequences and its applications.
|
| |
BMC Evol Biol,
4,
33.
|
 |
|
|
|
|
 |
M.Singleton,
M.Isupov,
and
J.Littlechild
(1999).
X-ray structure of pyrrolidone carboxyl peptidase from the hyperthermophilic archaeon Thermococcus litoralis.
|
| |
Structure,
7,
237-244.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Gerstein,
and
C.Chothia
(1996).
Packing at the protein-water interface.
|
| |
Proc Natl Acad Sci U S A,
93,
10167-10172.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

