 |
PDBsum entry 2c4c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transport
|
 |
|
Title:
|
 |
Crystal structure of the NADPH-treated monooxygenase domain of mical
|
|
Structure:
|
 |
Nedd9-interacting protein with calponin homology and lim domains. Chain: a, b. Synonym: mical, molecule interacting with casl protein 1. Engineered: yes
|
|
Source:
|
 |
Mus musculus. Mouse. Organism_taxid: 10090. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.90Å
|
R-factor:
|
0.246
|
R-free:
|
0.294
|
|
|
Authors:
|
 |
C.Siebold,N.Berrow,T.S.Walter,K.Harlos,R.J.Owens,J.R.Terman, D.I.Stuart,A.L.Kolodkin,R.J.Pasterkamp,E.Y.Jones
|
Key ref:

|
 |
C.Siebold
et al.
(2005).
High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule.
Proc Natl Acad Sci U S A,
102,
16836-16841.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
18-Oct-05
|
Release date:
|
26-Oct-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8VDP3
(MICA1_MOUSE) -
[F-actin]-monooxygenase MICAL1 from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1048 a.a.
476 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.14.13.225
- F-actin monooxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionyl-[F-actin] + NADPH + O2 + H+ = L-methionyl-(R)-S-oxide- [F-actin] + NADP+ + H2O
|
 |
 |
 |
 |
 |
L-methionyl-[F-actin]
|
+
|
 NADPH
NADPH
|
+
|
O2
Bound ligand (Het Group name = )
matches with 71.19% similarity
|
+
|
H(+)
|
=
|
L-methionyl-(R)-S-oxide- [F-actin]
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.6.3.1
- NAD(P)H oxidase (H2O2-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
NADPH + O2 + H+ = H2O2 + NADP+
|
|
2.
|
NADH + O2 + H+ = H2O2 + NAD+
|
|
 |
 |
 |
 |
 |
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
 H2O2
H2O2
|
+
|
 NADP(+)
NADP(+)
|
|
 |
 |
 |
 |
 |
 NADH
NADH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
 H2O2
H2O2
|
+
|
 NAD(+)
NAD(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Ca(2+); FAD; Heme
|
 |
 |
 |
 |
 |
Ca(2+)
|
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
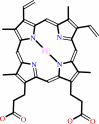 Heme
Heme
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
102:16836-16841
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule.
|
|
C.Siebold,
N.Berrow,
T.S.Walter,
K.Harlos,
R.J.Owens,
D.I.Stuart,
J.R.Terman,
A.L.Kolodkin,
R.J.Pasterkamp,
E.Y.Jones.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Semaphorins are extracellular cell guidance cues that govern cytoskeletal
dynamics during neuronal and vascular development. MICAL (molecule interacting
with CasL) is a multidomain cytosolic protein with a putative flavoprotein
monooxygenase (MO) region required for semaphorin-plexin repulsive axon
guidance. Here, we report the 1.45-A resolution crystal structure of the
FAD-containing MO domain of mouse MICAL-1 (residues 1-489). The topology most
closely resembles that of the NADPH-dependent flavoenzyme p-hydroxybenzoate
hydroxylase (PHBH). Comparison of structures before and after reaction with
NADPH reveals that, as in PHBH, the flavin ring can switch between two discrete
positions. In contrast with other MOs, this conformational switch is coupled
with the opening of a channel to the active site, suggestive of a protein
substrate. In support of this hypothesis, distinctive structural features
highlight putative protein-binding sites in suitable proximity to the active
site entrance. The unusual juxtaposition of this N-terminal MO (hydroxylase)
activity with the characteristics of a multiprotein-binding scaffold exhibited
by the C-terminal portion of the MICALs represents a unique combination of
functionality to mediate signaling.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Schematic representation of the FAD-apoprotein
interactions in mMICAL[489]. View on the si face of the flavin
with the FAD and interacting residues depicted as sticks [N,
blue; O, red; P, violet; S, yellow; C (protein), orange; C
(FAD), gray] and water molecules shown as cyan spheres. H bonds
are shown in green with lengths in Å. Red "eyelashes" show
hydrophobic interactions.
|
 |
Figure 4.
Fig. 4. Comparison of the reduced and oxidized forms of
mMICAL[489].(A and B) Superposition of the two forms. The FAD
molecules are drawn as balls and sticks (carbons of oxidized
mMICAL[489], cyan; carbons of reduced mMICAL[489], orange). The
main chain of the oxidized form is depicted as a ribbon. B is
rotated by 90° about the x axis relative to A. (C and D)
Coordination of the isoalloxazine ring in the oxidized (C) and
reduced (D) forms viewed from a common orientation. The
isoalloxazine ring and selected residues are depicted as sticks
(orange, carbon of reduced isoalloxazine; gray, protein carbon),
waters are shown as spheres, and H bonds are shown as yellow
dashes.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.J.Hung,
U.Yazdani,
J.Yoon,
H.Wu,
T.Yang,
N.Gupta,
Z.Huang,
W.J.van Berkel,
and
J.R.Terman
(2010).
Mical links semaphorins to F-actin disassembly.
|
| |
Nature,
463,
823-827.
|
 |
|
|
|
|
 |
F.Forneris,
R.Orru,
D.Bonivento,
L.R.Chiarelli,
and
A.Mattevi
(2009).
ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding.
|
| |
FEBS J,
276,
2833-2840.
|
 |
|
|
|
|
 |
K.Prochazkova,
L.A.Shuvalova,
G.Minasov,
Z.Voburka,
W.F.Anderson,
and
K.J.Satchell
(2009).
Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites.
|
| |
J Biol Chem,
284,
26557-26568.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Sun,
H.Zhou,
N.O.Binmadi,
and
J.R.Basile
(2009).
Hypoxia-inducible factor-1-mediated regulation of semaphorin 4D affects tumor growth and vascularity.
|
| |
J Biol Chem,
284,
32066-32074.
|
 |
|
|
|
|
 |
T.Senda,
M.Senda,
S.Kimura,
and
T.Ishida
(2009).
Redox control of protein conformation in flavoproteins.
|
| |
Antioxid Redox Signal,
11,
1741-1766.
|
 |
|
|
|
|
 |
E.F.Schmidt,
S.O.Shim,
and
S.M.Strittmatter
(2008).
Release of MICAL autoinhibition by semaphorin-plexin signaling promotes interaction with collapsin response mediator protein.
|
| |
J Neurosci,
28,
2287-2297.
|
 |
|
|
|
|
 |
E.F.Schmidt,
and
S.M.Strittmatter
(2007).
The CRMP family of proteins and their role in Sema3A signaling.
|
| |
Adv Exp Med Biol,
600,
1.
|
 |
|
|
|
|
 |
V.Joosten,
and
W.J.van Berkel
(2007).
Flavoenzymes.
|
| |
Curr Opin Chem Biol,
11,
195-202.
|
 |
|
|
|
|
 |
A.Geerlof,
J.Brown,
B.Coutard,
M.P.Egloff,
F.J.Enguita,
M.J.Fogg,
R.J.Gilbert,
M.R.Groves,
A.Haouz,
J.E.Nettleship,
P.Nordlund,
R.J.Owens,
M.Ruff,
S.Sainsbury,
D.I.Svergun,
and
M.Wilmanns
(2006).
The impact of protein characterization in structural proteomics.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1125-1136.
|
 |
|
|
|
|
 |
H.Sun,
H.Dai,
J.Zhang,
X.Jin,
S.Xiong,
J.Xu,
J.Wu,
and
Y.Shi
(2006).
Solution structure of calponin homology domain of Human MICAL-1.
|
| |
J Biomol NMR,
36,
295-300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Au,
N.S.Berrow,
E.Blagova,
I.W.Boucher,
M.P.Boyle,
J.A.Brannigan,
L.G.Carter,
T.Dierks,
G.Folkers,
R.Grenha,
K.Harlos,
R.Kaptein,
A.K.Kalliomaa,
V.M.Levdikov,
C.Meier,
N.Milioti,
O.Moroz,
A.Müller,
R.J.Owens,
N.Rzechorzek,
S.Sainsbury,
D.I.Stuart,
T.S.Walter,
D.G.Waterman,
A.J.Wilkinson,
K.S.Wilson,
N.Zaccai,
R.M.Esnouf,
and
M.J.Fogg
(2006).
Application of high-throughput technologies to a structural proteomics-type analysis of Bacillus anthracis.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1267-1275.
|
 |
|
|
|
|
 |
L.Banci,
I.Bertini,
S.Cusack,
R.N.de Jong,
U.Heinemann,
E.Y.Jones,
F.Kozielski,
K.Maskos,
A.Messerschmidt,
R.Owens,
A.Perrakis,
A.Poterszman,
G.Schneider,
C.Siebold,
I.Silman,
T.Sixma,
G.Stewart-Jones,
J.L.Sussman,
J.C.Thierry,
and
D.Moras
(2006).
First steps towards effective methods in exploiting high-throughput technologies for the determination of human protein structures of high biomedical value.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1208-1217.
|
 |
|
|
|
|
 |
L.De Colibus,
and
A.Mattevi
(2006).
New frontiers in structural flavoenzymology.
|
| |
Curr Opin Struct Biol,
16,
722-728.
|
 |
|
|
|
|
 |
M.J.Fogg,
P.Alzari,
M.Bahar,
I.Bertini,
J.M.Betton,
W.P.Burmeister,
C.Cambillau,
B.Canard,
M.A.Corrondo,
M.Carrondo,
M.Coll,
S.Daenke,
O.Dym,
M.P.Egloff,
F.J.Enguita,
A.Geerlof,
A.Haouz,
T.A.Jones,
Q.Ma,
S.N.Manicka,
M.Migliardi,
P.Nordlund,
R.J.Owens,
Y.Peleg,
G.Schneider,
R.Schnell,
D.I.Stuart,
N.Tarbouriech,
T.Unge,
A.J.Wilkinson,
M.Wilmanns,
K.S.Wilson,
O.Zimhony,
and
J.M.Grimes
(2006).
Application of the use of high-throughput technologies to the determination of protein structures of bacterial and viral pathogens.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1196-1207.
|
 |
|
|
|
|
 |
N.S.Berrow,
K.Büssow,
B.Coutard,
J.Diprose,
M.Ekberg,
G.E.Folkers,
N.Levy,
V.Lieu,
R.J.Owens,
Y.Peleg,
C.Pinaglia,
S.Quevillon-Cheruel,
L.Salim,
C.Scheich,
R.Vincentelli,
and
D.Busso
(2006).
Recombinant protein expression and solubility screening in Escherichia coli: a comparative study.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1218-1226.
|
 |
|
|
|
|
 |
S.Albeck,
P.Alzari,
C.Andreini,
L.Banci,
I.M.Berry,
I.Bertini,
C.Cambillau,
B.Canard,
L.Carter,
S.X.Cohen,
J.M.Diprose,
O.Dym,
R.M.Esnouf,
C.Felder,
F.Ferron,
F.Guillemot,
R.Hamer,
M.Ben Jelloul,
R.A.Laskowski,
T.Laurent,
S.Longhi,
R.Lopez,
C.Luchinat,
H.Malet,
T.Mochel,
R.J.Morris,
L.Moulinier,
T.Oinn,
A.Pajon,
Y.Peleg,
A.Perrakis,
O.Poch,
J.Prilusky,
A.Rachedi,
R.Ripp,
A.Rosato,
I.Silman,
D.I.Stuart,
J.L.Sussman,
J.C.Thierry,
J.D.Thompson,
J.M.Thornton,
T.Unger,
B.Vaughan,
W.Vranken,
J.D.Watson,
G.Whamond,
and
K.Henrick
(2006).
SPINE bioinformatics and data-management aspects of high-throughput structural biology.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1184-1195.
|
 |
|
|
|
|
 |
T.S.Walter,
C.Meier,
R.Assenberg,
K.F.Au,
J.Ren,
A.Verma,
J.E.Nettleship,
R.J.Owens,
D.I.Stuart,
and
J.M.Grimes
(2006).
Lysine methylation as a routine rescue strategy for protein crystallization.
|
| |
Structure,
14,
1617-1622.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

