
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.1.14.18.1
- tyrosinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Melanin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-tyrosine + O2 = L-dopaquinone + H2O
|
|
2.
|
2 L-dopa + O2 = 2 L-dopaquinone + 2 H2O
|
|
 |
 |
 |
 |
 |
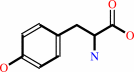 L-tyrosine
L-tyrosine
|
+
|
 O2
O2
|
=
|
L-dopaquinone
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
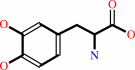 2
×
L-dopa
2
×
L-dopa
|
+
|
 O2
O2
|
=
|
2
×
L-dopaquinone
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:8981-8990
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis.
|
|
Y.Matoba,
T.Kumagai,
A.Yamamoto,
H.Yoshitsu,
M.Sugiyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
At high resolution, we determined the crystal structures of copper-bound and
metal-free tyrosinase in a complex with ORF378 designated as a "caddie" protein
because it assists with transportation of two CuII ions into the tyrosinase
catalytic center. These structures suggest that the caddie protein covers the
hydrophobic molecular surface of tyrosinase and interferes with the binding of a
substrate tyrosine to the catalytic site of tyrosinase. The caddie protein,
which consists of one six-strandedbeta-sheet and one alpha-helix, has no
similarity with all proteins deposited into the Protein Data Bank. Although
tyrosinase and catechol oxidase are classified into the type 3 copper protein
family, the latter enzyme lacks monooxygenase activity. The difference in
catalytic activity is based on the structural observations that a large vacant
space is present just above the active center of tyrosinase and that one of the
six His ligands for the two copper ions is highly flexible. These structural
characteristics of tyrosinase suggest that, in the reaction that catalyzes the
ortho-hydroxylation of monophenol, one of the two Cu(II) ions is coordinated by
the peroxide-originated oxygen bound to the substrate. Our crystallographic
study shows evidence that the tyrosinase active center formed by dinuclear
coppers is flexible during catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
FIGURE 5. Structural similarity. A, stereo representation
of the superposition of tyrosinase and structurally homologous
proteins, potato catechol oxidase (Protein Data Bank code 1BT1)
and octopus hemocyanin (Protein Data Bank code 1JS8). Red, blue,
and green indicate the backbone traces of tyrosinase, catechol
oxidase, and hemocyanin, respectively. The yellow sphere
indicates the two copper ions in the catalytic center. To
emphasize similarity, the C-terminal domain of hemocyanin is
omitted from the figure. B, stereo view of the superposition of
ORF378 and the SH2 domain in the growth factor-bound protein 2
(Protein Data Bank code 1GRI). Red and blue indicate the
backbone trace of ORF378 and the SH2 domain in the growth
factor-bound protein 2, respectively.
|
 |
Figure 9.
FIGURE 9. Structure-based catalytic mechanism of
tyrosinase. The oxy form of tyrosinase catalyzes the conversion
of monophenol to the corresponding quinone through the
ortho-diphenol formation. In this scheme, His^54 is released
from the Cu^A site, resulting in the formation of the bidentate
intermediate. The met and oxy forms of tyrosinase can catalyze
the conversion of ortho-diphenol to the corresponding quinone.
This reaction should progress similarly to that of catechol
oxidase.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
8981-8990)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Jaenicke,
K.Büchler,
H.Decker,
J.Markl,
and
G.F.Schröder
(2011).
The refined structure of functional unit h of keyhole limpet hemocyanin (KLH1-h) reveals disulfide bridges.
|
| |
IUBMB Life,
63,
183-187.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.G.Mutti,
M.Gullotti,
L.Casella,
L.Santagostini,
R.Pagliarin,
K.K.Andersson,
M.F.Iozzi,
and
G.Zoppellaro
(2011).
A new chiral, poly-imidazole N8-ligand and the related di- and tri-copper(II) complexes: synthesis, theoretical modelling, spectroscopic properties, and biomimetic stereoselective oxidations.
|
| |
Dalton Trans,
40,
5436-5457.
|
 |
|
|
|
|
 |
H.M.Wang,
C.Y.Chen,
and
Z.H.Wen
(2011).
Identifying melanogenesis inhibitors from Cinnamomum subavenium with in vitro and in vivo screening systems by targeting the human tyrosinase.
|
| |
Exp Dermatol,
20,
242-248.
|
 |
|
|
|
|
 |
Y.J.Choi,
K.B.Cho,
M.Kubo,
T.Ogura,
K.D.Karlin,
J.Cho,
and
W.Nam
(2011).
Spectroscopic and computational characterization of Cu(II)-OOR (R = H or cumyl) complexes bearing a Me(6)-tren ligand.
|
| |
Dalton Trans,
40,
2234-2241.
|
 |
|
|
|
|
 |
Y.Piao,
Z.Jin,
D.Lee,
H.J.Lee,
H.B.Na,
T.Hyeon,
M.K.Oh,
J.Kim,
and
H.S.Kim
(2011).
Sensitive and high-fidelity electrochemical immunoassay using carbon nanotubes coated with enzymes and magnetic nanoparticles.
|
| |
Biosens Bioelectron,
26,
3192-3199.
|
 |
|
|
|
|
 |
A.de la Lande,
J.Maddaluno,
O.Parisel,
T.A.Darden,
and
J.P.Piquemal
(2010).
Study of the docking of competitive inhibitors at a model of tyrosinase active site: insights from joint broken-symmetry/Spin-Flip DFT computations and ELF topological analysis.
|
| |
Interdiscip Sci,
2,
3.
|
 |
|
|
|
|
 |
C.Gasparetti,
G.Faccio,
M.Arvas,
J.Buchert,
M.Saloheimo,
and
K.Kruus
(2010).
Discovery of a new tyrosinase-like enzyme family lacking a C-terminally processed domain: production and characterization of an Aspergillus oryzae catechol oxidase.
|
| |
Appl Microbiol Biotechnol,
86,
213-226.
|
 |
|
|
|
|
 |
C.J.Vavricka,
B.M.Christensen,
and
J.Li
(2010).
Melanization in living organisms: a perspective of species evolution.
|
| |
Protein Cell,
1,
830-841.
|
 |
|
|
|
|
 |
C.Núñez,
R.Bastida,
A.Macías,
L.Valencia,
N.I.Neuman,
A.C.Rizzi,
C.D.Brondino,
P.J.González,
J.L.Capelo,
and
C.Lodeiro
(2010).
Structural, MALDI-TOF-MS, magnetic and spectroscopic studies of new dinuclear copper(II), cobalt(II) and zinc(II) complexes containing a biomimicking μ-OH bridge.
|
| |
Dalton Trans,
39,
11654-11663.
|
 |
|
|
|
|
 |
E.Jaenicke,
K.Büchler,
J.Markl,
H.Decker,
and
T.R.Barends
(2010).
Cupredoxin-like domains in haemocyanins.
|
| |
Biochem J,
426,
373-378.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Garcia-Molina,
J.L.Munoz-Munoz,
M.Garcia-Molina,
P.A.Garcia-Ruiz,
J.Tudela,
F.García-Cánovas,
and
J.N.Rodriguez-Lopez
(2010).
Melanogenesis inhibition due to NADH.
|
| |
Biosci Biotechnol Biochem,
74,
1777-1787.
|
 |
|
|
|
|
 |
J.A.Worrall,
and
E.Vijgenboom
(2010).
Copper mining in Streptomyces: enzymes, natural products and development.
|
| |
Nat Prod Rep,
27,
742-756.
|
 |
|
|
|
|
 |
J.L.Muñoz-Muñoz,
F.Garcia-Molina,
R.Varon,
P.A.Garcia-Ruíz,
J.Tudela,
F.Garcia-Cánovas,
and
J.N.Rodríguez-López
(2010).
Suicide inactivation of the diphenolase and monophenolase activities of tyrosinase.
|
| |
IUBMB Life,
62,
539-547.
|
 |
|
|
|
|
 |
M.Fairhead,
and
L.Thöny-Meyer
(2010).
Role of the C-terminal extension in a bacterial tyrosinase.
|
| |
FEBS J,
277,
2083-2095.
|
 |
|
|
|
|
 |
N.Fujieda,
A.Yakiyama,
and
S.Itoh
(2010).
Catalytic oxygenation of phenols by arthropod hemocyanin, an oxygen carrier protein, from Portunus trituberculatus.
|
| |
Dalton Trans,
39,
3083-3092.
|
 |
|
|
|
|
 |
R.D.Kersten,
and
P.C.Dorrestein
(2010).
Metalloenzymes: Natural product nitrosation.
|
| |
Nat Chem Biol,
6,
636-637.
|
 |
|
|
|
|
 |
R.J.Deeth,
and
C.Diedrich
(2010).
Structural and mechanistic insights into the oxy form of tyrosinase from molecular dynamics simulations.
|
| |
J Biol Inorg Chem,
15,
117-129.
|
 |
|
|
|
|
 |
R.K.Das,
B.Saha,
S.M.Rahaman,
and
J.K.Bera
(2010).
Bimetallic catalysis involving dipalladium(I) and diruthenium(I) complexes.
|
| |
Chemistry,
16,
14459-14468.
|
 |
|
|
|
|
 |
Y.J.Guo,
Z.Z.Pan,
C.Q.Chen,
Y.H.Hu,
F.J.Liu,
Y.Shi,
J.H.Yan,
and
Q.X.Chen
(2010).
Inhibitory effects of fatty acids on the activity of mushroom tyrosinase.
|
| |
Appl Biochem Biotechnol,
162,
1564-1573.
|
 |
|
|
|
|
 |
A.Spada,
S.Palavicini,
E.Monzani,
L.Bubacco,
and
L.Casella
(2009).
Trapping tyrosinase key active intermediate under turnover.
|
| |
Dalton Trans,
(),
6468-6471.
|
 |
|
|
|
|
 |
B.T.Op't Holt,
M.A.Vance,
L.M.Mirica,
D.E.Heppner,
T.D.Stack,
and
E.I.Solomon
(2009).
Reaction coordinate of a functional model of tyrosinase: spectroscopic and computational characterization.
|
| |
J Am Chem Soc,
131,
6421-6438.
|
 |
|
|
|
|
 |
C.Olivares,
and
F.Solano
(2009).
New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins.
|
| |
Pigment Cell Melanoma Res,
22,
750-760.
|
 |
|
|
|
|
 |
H.Y.Yang,
and
C.W.Chen
(2009).
Extracellular and intracellular polyphenol oxidases cause opposite effects on sensitivity of streptomyces to phenolics: a case of double-edged sword.
|
| |
PLoS One,
4,
e7462.
|
 |
|
|
|
|
 |
J.Yoon,
S.Fujii,
and
E.I.Solomon
(2009).
Geometric and electronic structure differences between the type 3 copper sites of the multicopper oxidases and hemocyanin/tyrosinase.
|
| |
Proc Natl Acad Sci U S A,
106,
6585-6590.
|
 |
|
|
|
|
 |
M.Güell,
J.M.Luis,
M.Solà,
and
P.E.Siegbahn
(2009).
Theoretical study of the hydroxylation of phenolates by the Cu(2)O (2)(N,N'-dimethylethylenediamine) (2) (2+) complex.
|
| |
J Biol Inorg Chem,
14,
229-242.
|
 |
|
|
|
|
 |
S.Herres-Pawlis,
P.Verma,
R.Haase,
P.Kang,
C.T.Lyons,
E.C.Wasinger,
U.Flörke,
G.Henkel,
and
T.D.Stack
(2009).
Phenolate hydroxylation in a bis(mu-oxo)dicopper(III) complex: lessons from the guanidine/amine series.
|
| |
J Am Chem Soc,
131,
1154-1169.
|
 |
|
|
|
|
 |
S.Herres-Pawlis,
S.Binder,
A.Eich,
R.Haase,
B.Schulz,
G.Wellenreuther,
G.Henkel,
M.Rübhausen,
and
W.Meyer-Klaucke
(2009).
Stabilisation of a highly reactive bis(mu-oxo)dicopper(III) species at room temperature by electronic and steric constraint of an unconventional nitrogen donor ligand.
|
| |
Chemistry,
15,
8678-8682.
|
 |
|
|
|
|
 |
T.S.Chang
(2009).
An updated review of tyrosinase inhibitors.
|
| |
Int J Mol Sci,
10,
2440-2475.
|
 |
|
|
|
|
 |
Y.Cong,
Q.Zhang,
D.Woolford,
T.Schweikardt,
H.Khant,
M.Dougherty,
S.J.Ludtke,
W.Chiu,
and
H.Decker
(2009).
Structural mechanism of SDS-induced enzyme activity of scorpion hemocyanin revealed by electron cryomicroscopy.
|
| |
Structure,
17,
749-758.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Li,
Y.Wang,
H.Jiang,
and
J.Deng
(2009).
Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
|
| |
Proc Natl Acad Sci U S A,
106,
17002-17006.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Brack,
N.Hellmann,
and
H.Decker
(2008).
Kinetic properties of hexameric tyrosinase from the crustacean Palinurus elephas.
|
| |
Photochem Photobiol,
84,
692-699.
|
 |
|
|
|
|
 |
A.Prokofieva,
A.I.Prikhod'ko,
S.Dechert,
and
F.Meyer
(2008).
Selective benzylic C-C coupling catalyzed by a bioinspired dicopper complex.
|
| |
Chem Commun (Camb),
(),
1005-1007.
|
 |
|
|
|
|
 |
E.J.Land,
C.A.Ramsden,
P.A.Riley,
and
M.R.Stratford
(2008).
Evidence consistent with the requirement of cresolase activity for suicide inactivation of tyrosinase.
|
| |
Tohoku J Exp Med,
216,
231-238.
|
 |
|
|
|
|
 |
E.Jaenicke,
and
H.Decker
(2008).
Kinetic properties of catecholoxidase activity of tarantula hemocyanin.
|
| |
FEBS J,
275,
1518-1528.
|
 |
|
|
|
|
 |
F.G.Mutti,
R.Pievo,
M.Sgobba,
M.Gullotti,
and
L.Santagostini
(2008).
Biomimetic modeling of copper complexes: a study of enantioselective catalytic oxidation on d-(+)-catechin and L-( - )-epicatechin with copper complexes.
|
| |
Bioinorg Chem Appl,
(),
762029.
|
 |
|
|
|
|
 |
L.Q.Hatcher,
and
J.D.Simon
(2008).
Ultra-low temperature oxidation of 5,6-dihydroxyindole: a novel approach to study synthetic melanogenesis.
|
| |
Photochem Photobiol,
84,
608-612.
|
 |
|
|
|
|
 |
L.Que,
and
W.B.Tolman
(2008).
Biologically inspired oxidation catalysis.
|
| |
Nature,
455,
333-340.
|
 |
|
|
|
|
 |
S.Hirota,
T.Kawahara,
M.Beltramini,
P.Di Muro,
R.S.Magliozzo,
J.Peisach,
L.S.Powers,
N.Tanaka,
S.Nagao,
and
L.Bubacco
(2008).
Molecular basis of the Bohr effect in arthropod hemocyanin.
|
| |
J Biol Chem,
283,
31941-31948.
|
 |
|
|
|
|
 |
A.Company,
L.Gómez,
R.Mas-Ballesté,
I.V.Korendovych,
X.Ribas,
A.Poater,
T.Parella,
X.Fontrodona,
J.Benet-Buchholz,
M.Solà,
L.Que,
E.V.Rybak-Akimova,
and
M.Costas
(2007).
Fast O2 binding at dicopper complexes containing Schiff-base dinucleating ligands.
|
| |
Inorg Chem,
46,
4997-5012.
|
 |
|
|
|
|
 |
A.S.Hakemian,
and
A.C.Rosenzweig
(2007).
The biochemistry of methane oxidation.
|
| |
Annu Rev Biochem,
76,
223-241.
|
 |
|
|
|
|
 |
A.de la Lande,
V.Moliner,
and
O.Parisel
(2007).
Singlet-triplet gaps in large multireference systems: spin-flip-driven alternatives for bioinorganic modeling.
|
| |
J Chem Phys,
126,
035102.
|
 |
|
|
|
|
 |
C.B.Xue,
W.C.Luo,
L.Jiang,
X.Y.Xie,
T.Xiao,
and
L.Yan
(2007).
Inhibition kinetics of cabbage butterfly (Pieris rapae L.) larvae phenoloxidase activity by 3-hydroxy-4-methoxybenzaldehyde thiosemicarbazone.
|
| |
Appl Biochem Biotechnol,
143,
101-114.
|
 |
|
|
|
|
 |
E.J.Land,
C.A.Ramsden,
and
P.A.Riley
(2007).
The mechanism of suicide-inactivation of tyrosinase: a substrate structure investigation.
|
| |
Tohoku J Exp Med,
212,
341-348.
|
 |
|
|
|
|
 |
H.B.Albada,
F.Soulimani,
B.M.Weckhuysen,
and
R.M.Liskamp
(2007).
Scaffolded amino acids as a close structural mimic of type-3 copper binding sites.
|
| |
Chem Commun (Camb),
(),
4895-4897.
|
 |
|
|
|
|
 |
J.Yoon,
and
E.I.Solomon
(2007).
Electronic structure of the peroxy intermediate and its correlation to the native intermediate in the multicopper oxidases: insights into the reductive cleavage of the o-o bond.
|
| |
J Am Chem Soc,
129,
13127-13136.
|
 |
|
|
|
|
 |
K.Born,
P.Comba,
A.Daubinet,
A.Fuchs,
and
H.Wadepohl
(2007).
Catecholase activity of dicopper(II)-bispidine complexes: stabilities and structures of intermediates, kinetics and reaction mechanism.
|
| |
J Biol Inorg Chem,
12,
36-48.
|
 |
|
|
|
|
 |
M.Güell,
and
P.E.Siegbahn
(2007).
Theoretical study of the catalytic mechanism of catechol oxidase.
|
| |
J Biol Inorg Chem,
12,
1251-1264.
|
 |
|
|
|
|
 |
P.Nicholls
(2007).
The oxygenase-peroxidase theory of Bach and Chodat and its modern equivalents: change and permanence in scientific thinking as shown by our understanding of the roles of water, peroxide, and oxygen in the functioning of redox enzymes.
|
| |
Biochemistry (Mosc),
72,
1039-1046.
|
 |
|
|
|
|
 |
S.R.Kanade,
V.L.Suhas,
N.Chandra,
and
L.R.Gowda
(2007).
Functional interaction of diphenols with polyphenol oxidase. Molecular determinants of substrate/inhibitor specificity.
|
| |
FEBS J,
274,
4177-4187.
|
 |
|
|
|
|
 |
S.Takahashi,
H.Iwai,
K.Kosaka,
T.Miyazaki,
Y.Osanai,
N.Arao,
K.Tanaka,
K.Nagai,
and
A.Nakagawa
(2007).
Byelyankacin: a novel melanogenesis inhibitor produced by Enterobacter sp. B20.
|
| |
J Antibiot (Tokyo),
60,
717-720.
|
 |
|
|
|
|
 |
Y.Kawamura-Konishi,
M.Tsuji,
S.Hatana,
M.Asanuma,
D.Kakuta,
T.Kawano,
E.B.Mukouyama,
H.Goto,
and
H.Suzuki
(2007).
Purification, characterization, and molecular cloning of tyrosinase from Pholiota nameko.
|
| |
Biosci Biotechnol Biochem,
71,
1752-1760.
|
 |
|
|
|
|
 |
A.C.Rosenzweig,
and
M.H.Sazinsky
(2006).
Structural insights into dioxygen-activating copper enzymes.
|
| |
Curr Opin Struct Biol,
16,
729-735.
|
 |
|
|
|
|
 |
A.Company,
D.Lamata,
A.Poater,
M.Solà,
E.V.Rybak-Akimova,
L.Que,
X.Fontrodona,
T.Parella,
A.Llobet,
and
M.Costas
(2006).
O2 chemistry of dicopper complexes with alkyltriamine ligands. Comparing synergistic effects on O2 binding.
|
| |
Inorg Chem,
45,
5239-5241.
|
 |
|
|
|
|
 |
E.Selinheimo,
M.Saloheimo,
E.Ahola,
A.Westerholm-Parvinen,
N.Kalkkinen,
J.Buchert,
and
K.Kruus
(2006).
Production and characterization of a secreted, C-terminally processed tyrosinase from the filamentous fungus Trichoderma reesei.
|
| |
FEBS J,
273,
4322-4335.
|
 |
|
|
|
|
 |
H.Decker,
T.Schweikardt,
and
F.Tuczek
(2006).
The first crystal structure of tyrosinase: all questions answered?
|
| |
Angew Chem Int Ed Engl,
45,
4546-4550.
|
 |
|
|
|
|
 |
S.Itoh,
and
Y.Tachi
(2006).
Structure and O2-reactivity of copper(I) complexes supported by pyridylalkylamine ligands.
|
| |
Dalton Trans,
(),
4531-4538.
|
 |
|
|
|
|
 |
X.Ottenwaelder,
D.J.Rudd,
M.C.Corbett,
K.O.Hodgson,
B.Hedman,
and
T.D.Stack
(2006).
Reversible O-O bond cleavage in copper-dioxygen isomers: impact of anion basicity.
|
| |
J Am Chem Soc,
128,
9268-9269.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

