 |
PDBsum entry 2a9c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2a9c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of r138q mutant of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site
|
|
Structure:
|
 |
Sulfite oxidase. Chain: a, b. Fragment: catalytic core domain and c terminal dimerization domain. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Gallus gallus. Chicken. Organism_taxid: 9031. Organ: liver. Organelle: mitochondrion. Cellular_location: mitochondrial intermembrane space. Gene: suox. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.51Å
|
R-factor:
|
0.160
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
E.Karakas,H.L.Wilson,T.N.Graf,S.Xiang,S.Jaramillo-Busquets, K.V.Rajagopalan,C.Kisker
|
Key ref:

|
 |
E.Karakas
et al.
(2005).
Structural insights into sulfite oxidase deficiency.
J Biol Chem,
280,
33506-33515.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Jul-05
|
Release date:
|
02-Aug-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07850
(SUOX_CHICK) -
Sulfite oxidase from Gallus gallus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
459 a.a.
359 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 7 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.3.1
- sulfite oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
sulfite + O2 + H2O = sulfate + H2O2
|
 |
 |
 |
 |
 |
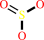 sulfite
sulfite
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
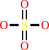 sulfate
sulfate
|
+
|
H2O2
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme; Mo cation
|
 |
 |
 |
 |
 |
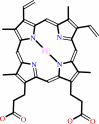 Heme
Heme
|
Mo cation
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:33506-33515
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into sulfite oxidase deficiency.
|
|
E.Karakas,
H.L.Wilson,
T.N.Graf,
S.Xiang,
S.Jaramillo-Busquets,
K.V.Rajagopalan,
C.Kisker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Sulfite oxidase deficiency is a lethal genetic disease that results from defects
either in the genes encoding proteins involved in molybdenum cofactor
biosynthesis or in the sulfite oxidase gene itself. Several point mutations in
the sulfite oxidase gene have been identified from patients suffering from this
disease worldwide. Although detailed biochemical analyses have been carried out
on these mutations, no structural data could be obtained because of problems in
crystallizing recombinant human and rat sulfite oxidases and the failure to
clone the chicken sulfite oxidase gene. We synthesized the gene for chicken
sulfite oxidase de novo, working backward from the amino acid sequence of the
native chicken liver enzyme by PCR amplification of a series of 72 overlapping
primers. The recombinant protein displayed the characteristic absorption
spectrum of sulfite oxidase and exhibited steady state and rapid kinetic
parameters comparable with those of the tissue-derived enzyme. We solved the
crystal structures of the wild type and the sulfite oxidase deficiency-causing
R138Q (R160Q in humans) variant of recombinant chicken sulfite oxidase in the
resting and sulfate-bound forms. Significant alterations in the
substrate-binding pocket were detected in the structure of the mutant, and a
comparison between the wild type and mutant protein revealed that the active
site residue Arg-450 adopts different conformations in the presence and absence
of bound sulfate. The size of the binding pocket is thereby considerably
reduced, and its position relative to the cofactor is shifted, causing an
increase in the distance of the sulfur atom of the bound sulfate to the
molybdenum.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIGURE 4. Superposition of the active sites of the sulfate
bound (yellow) and unbound rCSO (gray). Only the Moco of the
sulfate free structure is shown. The molybdenum is shown in
green, and the water/hydroxo ligand is shown as a red sphere.
Hydrogen bonds between the sulfate and Arg-450 are shown with
dashed lines.
|
 |
Figure 8.
FIGURE 8. Proposed reaction mechanism for sulfite oxidase.
Schematic presentation of the reaction mechanism for the
oxidation of sulfite to sulfate. A, oxidized Mo center. B, the
Mo(IV)-sulfate adduct formed after an attack of sulfite on the
equatorial oxo group. C, the free reduced Mo(IV) center after
release of sulfate.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
33506-33515)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.V.Bernhardt
(2011).
Exploiting the versatility and selectivity of Mo enzymes with electrochemistry.
|
| |
Chem Commun (Camb),
47,
1663-1673.
|
 |
|
|
|
|
 |
A.V.Astashkin,
E.L.Klein,
D.Ganyushin,
K.Johnson-Winters,
F.Neese,
U.Kappler,
and
J.H.Enemark
(2009).
Exchangeable oxygens in the vicinity of the molybdenum center of the high-pH form of sulfite oxidase and sulfite dehydrogenase.
|
| |
Phys Chem Chem Phys,
11,
6733-6742.
|
 |
|
|
|
|
 |
E.L.Klein,
A.V.Astashkin,
D.Ganyushin,
C.Riplinger,
K.Johnson-Winters,
F.Neese,
and
J.H.Enemark
(2009).
Direct detection and characterization of chloride in the active site of the low-pH form of sulfite oxidase using electron spin echo envelope modulation spectroscopy, isotopic labeling, and density functional theory calculations.
|
| |
Inorg Chem,
48,
4743-4752.
|
 |
|
|
|
|
 |
M.J.Romão
(2009).
Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview.
|
| |
Dalton Trans,
(),
4053-4068.
|
 |
|
|
|
|
 |
S.Bailey,
T.Rapson,
K.Johnson-Winters,
A.V.Astashkin,
J.H.Enemark,
and
U.Kappler
(2009).
Molecular basis for enzymatic sulfite oxidation: how three conserved active site residues shape enzyme activity.
|
| |
J Biol Chem,
284,
2053-2063.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Emesh,
T.D.Rapson,
A.Rajapakshe,
U.Kappler,
P.V.Bernhardt,
G.Tollin,
and
J.H.Enemark
(2009).
Intramolecular electron transfer in sulfite-oxidizing enzymes: elucidating the role of a conserved active site arginine.
|
| |
Biochemistry,
48,
2156-2163.
|
 |
|
|
|
|
 |
A.V.Astashkin,
K.Johnson-Winters,
E.L.Klein,
C.Feng,
H.L.Wilson,
K.V.Rajagopalan,
A.M.Raitsimring,
and
J.H.Enemark
(2008).
Structural studies of the molybdenum center of the pathogenic R160Q mutant of human sulfite oxidase by pulsed EPR spectroscopy and 17O and 33S labeling.
|
| |
J Am Chem Soc,
130,
8471-8480.
|
 |
|
|
|
|
 |
C.J.Doonan,
H.L.Wilson,
B.Bennett,
R.C.Prince,
K.V.Rajagopalan,
and
G.N.George
(2008).
MoV electron paramagnetic resonance of sulfite oxidase revisited: the low-pH chloride signal.
|
| |
Inorg Chem,
47,
2033-2038.
|
 |
|
|
|
|
 |
G.J.Workun,
K.Moquin,
R.A.Rothery,
and
J.H.Weiner
(2008).
Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold.
|
| |
Microbiol Mol Biol Rev,
72,
228.
|
 |
|
|
|
|
 |
R.S.Sengar,
J.J.Miller,
and
P.Basu
(2008).
Design, syntheses, and characterization of dioxo-molybdenum(VI) complexes with thiolate ligands: effects of intraligand NH...S hydrogen bonding.
|
| |
Dalton Trans,
(),
2569-2577.
|
 |
|
|
|
|
 |
R.S.Sengar,
V.N.Nemykin,
and
P.Basu
(2008).
Synthesis, electrochemistry, geometric and electronic structure of oxo-molybdenum compounds involved in an oxygen atom transferring system.
|
| |
J Inorg Biochem,
102,
748-756.
|
 |
|
|
|
|
 |
C.Feng,
G.Tollin,
and
J.H.Enemark
(2007).
Sulfite oxidizing enzymes.
|
| |
Biochim Biophys Acta,
1774,
527-539.
|
 |
|
|
|
|
 |
J.H.Enemark,
A.V.Astashkin,
and
A.M.Raitsimring
(2006).
Investigation of the coordination structures of the molybdenum(v) sites of sulfite oxidizing enzymes by pulsed EPR spectroscopy.
|
| |
Dalton Trans,
(),
3501-3514.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

