 |
PDBsum entry 2d06

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Human sult1a1 complexed with pap and estradiol
|
|
Structure:
|
 |
Sulfotransferase 1a1. Chain: a, b. Synonym: sult1a1, phenol sulfotransferase 1, aryl sulfotransferase 1, phenol-sulfating phenol sulfotransferase 1, p- pst 1, thermostable phenol sulfotransferase, ts-pst, hast1/hast2, st1a3. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Tissue: liver. Gene: sult1a1. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.217
|
R-free:
|
0.283
|
|
|
Authors:
|
 |
N.U.Gamage,S.Tsvetanov,R.G.Duggleby,M.E.Mcmanus,J.L.Martin
|
Key ref:

|
 |
N.U.Gamage
et al.
(2005).
The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi-ring substrates.
J Biol Chem,
280,
41482-41486.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
25-Jul-05
|
Release date:
|
25-Oct-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P50225
(ST1A1_HUMAN) -
Sulfotransferase 1A1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
295 a.a.
288 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.8.2.1
- aryl sulfotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phenol + 3'-phosphoadenylyl sulfate = an aryl sulfate + adenosine 3',5'-bisphosphate + H+
|
 |
 |
 |
 |
 |
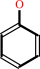 phenol
phenol
|
+
|
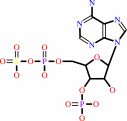 3'-phosphoadenylyl sulfate
3'-phosphoadenylyl sulfate
|
=
|
aryl sulfate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
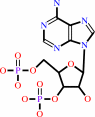 adenosine 3',5'-bisphosphate
adenosine 3',5'-bisphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:41482-41486
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi-ring substrates.
|
|
N.U.Gamage,
S.Tsvetanov,
R.G.Duggleby,
M.E.McManus,
J.L.Martin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human SULT1A1 belongs to the supergene family of sulfotransferases (SULTs)
involved in the sulfonation of xeno- and endobiotics. The enzyme is also one of
the SULTs responsible for metabolic activation of mutagenic and carcinogenic
compounds and therefore is implicated in various cancer forms. Further, it is
not well understood how substrate inhibition takes place with rigid fused
multiring substrates such as 17beta-estradiol (E2) at high substrate
concentrations when subcellular fractions or recombinant enzymes are used. To
investigate how estradiol binds to SULT1A1, we co-crystallized SULT1A1 with
sulfated estradiol and the cofactor product, PAP (3'-phosphoadenosine
5'-phosphate). The crystal structure of SULT1A1 that we present here has PAP and
one molecule of E2 bound in a nonproductive mode in the active site. The
structure reveals how the SULT1A1 binding site undergoes conformational changes
to accept fused ring substrates such as steroids. In agreement with previous
reports, the enzyme shows partial substrate inhibition at high concentrations of
E2. A model to explain these kinetics is developed based on the formation of an
enzyme x PAP x E2 dead-end complex during catalysis. This model provides a very
curve. This dead-end
complex is proposed to be that described by the observed structure, where E2 is
bound in a nonproductive mode.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIGURE 2. Comparison of fused ring substrate binding in
SULT1A1, mouse SULT1E1, and human SULT2A1. A, stereoview of E2
binding in the SULT1A1 active site. The noncatalytic substrate
binding mode of SULT1A1 is shown in orange. The O-3 hydroxyl of
E2 can form hydrogen bonds (dotted lines) to PAP and Lys48 (not
shown for clarity). The proposed E2 catalytic orientation
(green) was modeled using GOLD. For comparison, the E2 binding
mode from mouse SULT1E1 is shown (light blue). B,
superimposition of C  traces of
SULT1A1·PAP·pNP (pink) and
SULT1A1·PAP·E2 (green). E2 is shown in orange. A
loop (residues 84-90) that moves to accommodate E2 in the active
site is highlighted in dark blue and indicated by an arrow. traces of
SULT1A1·PAP·pNP (pink) and
SULT1A1·PAP·E2 (green). E2 is shown in orange. A
loop (residues 84-90) that moves to accommodate E2 in the active
site is highlighted in dark blue and indicated by an arrow.
|
 |
Figure 4.
FIGURE 4. Stereoview comparing the binding modes of E2
(orange) bound to SULT1A1 and DHEA bound to SULT2A1. The
productive or catalytic orientation of DHEA is shown in green,
and the nonproductive orientation is in black. PAP (light blue)
and the catalytic His108 (gray) of SULT1A1 are shown in a
ball-and-stick representation.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
41482-41486)
copyright 2005.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.B.Cole,
G.Keum,
J.Liu,
G.W.Small,
N.Satyamurthy,
V.Kepe,
and
J.R.Barrio
(2010).
Specific estrogen sulfotransferase (SULT1E1) substrates and molecular imaging probe candidates.
|
| |
Proc Natl Acad Sci U S A,
107,
6222-6227.
|
 |
|
|
|
|
 |
O.Khersonsky,
and
D.S.Tawfik
(2010).
Enzyme promiscuity: a mechanistic and evolutionary perspective.
|
| |
Annu Rev Biochem,
79,
471-505.
|
 |
|
|
|
|
 |
J.R.Pasqualini
(2009).
Estrogen sulfotransferases in breast and endometrial cancers.
|
| |
Ann N Y Acad Sci,
1155,
88-98.
|
 |
|
|
|
|
 |
J.Ziegler,
W.Brandt,
R.Geissler,
and
P.J.Facchini
(2009).
Removal of substrate inhibition and increase in maximal velocity in the short chain dehydrogenase/reductase salutaridine reductase involved in morphine biosynthesis.
|
| |
J Biol Chem,
284,
26758-26767.
|
 |
|
|
|
|
 |
G.E.Townsend,
and
D.H.Keating
(2008).
Identification and characterization of KpsS, a novel polysaccharide sulphotransferase in Mesorhizobium loti.
|
| |
Mol Microbiol,
68,
1149-1164.
|
 |
|
|
|
|
 |
A.Allali-Hassani,
P.W.Pan,
L.Dombrovski,
R.Najmanovich,
W.Tempel,
A.Dong,
P.Loppnau,
F.Martin,
J.Thornton,
J.Thonton,
A.M.Edwards,
A.Bochkarev,
A.N.Plotnikov,
M.Vedadi,
and
C.H.Arrowsmith
(2007).
Structural and chemical profiling of the human cytosolic sulfotransferases.
|
| |
PLoS Biol,
5,
e97.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Marsolais,
J.Boyd,
Y.Paredes,
A.M.Schinas,
M.Garcia,
S.Elzein,
and
L.Varin
(2007).
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760).
|
| |
Planta,
225,
1233-1244.
|
 |
|
|
|
|
 |
N.Hempel,
N.Gamage,
J.L.Martin,
and
M.E.McManus
(2007).
Human cytosolic sulfotransferase SULT1A1.
|
| |
Int J Biochem Cell Biol,
39,
685-689.
|
 |
|
|
|
|
 |
L.Dombrovski,
A.Dong,
A.Bochkarev,
and
A.N.Plotnikov
(2006).
Crystal structures of human sulfotransferases SULT1B1 and SULT1C1 complexed with the cofactor product adenosine-3'- 5'-diphosphate (PAP).
|
| |
Proteins,
64,
1091-1094.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

