 |
PDBsum entry 1x9d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of human class i alpha-1,2-mannosidase in complex with thio-disaccharide substrate analogue
|
|
Structure:
|
 |
Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha- mannosidase. Chain: a. Fragment: residues 243-699. Synonym: er alpha-1,2-mannosidase, mannosidase alpha class 1b member 1, man9glcnac2-specific processing alpha-mannosidase, unq747/pro1477. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: man1b1. Expressed in: pichia pastoris. Expression_system_taxid: 4922
|
|
Resolution:
|
 |
|
1.41Å
|
R-factor:
|
0.146
|
R-free:
|
0.162
|
|
|
Authors:
|
 |
K.Karaveg,W.Tempel,Z.J.Liu,A.Siriwardena,K.W.Moremen,B.C.Wang
|
Key ref:

|
 |
K.Karaveg
et al.
(2005).
Mechanism of class 1 (glycosylhydrolase family 47) {alpha}-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control.
J Biol Chem,
280,
16197-16207.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
20-Aug-04
|
Release date:
|
22-Feb-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9UKM7
(MA1B1_HUMAN) -
Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
699 a.a.
452 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.113
- mannosyl-oligosaccharide 1,2-alpha-mannosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N4-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 9A1,2,3B1,2,3) + 4 H2O = N4-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 5A1,2) + 4 beta-D-mannose
|
|
2.
|
N4-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->6)]-alpha-D-Man- (1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 8A1,2,3B1,3) + 3 H2O = N4-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man-(1->6)]-alpha- D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 5A1,2) + 3 beta-D-mannose.CC -!- This family of mammalian enzymes, located in the Golgi system,
|
|
 |
 |
 |
 |
 |
N(4)-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 9A1,2,3B1,2,3)
|
+
|
4
×
H2O
|
=
|
N(4)-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 5A1,2)
Bound ligand (Het Group name = )
matches with 78.57% similarity
|
+
|
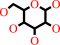 4
×
beta-D-mannose
4
×
beta-D-mannose
|
|
 |
 |
 |
 |
 |
N(4)-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->6)]-alpha-D-Man- (1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 8A1,2,3B1,3)
|
+
|
3
×
H2O
|
=
|
N(4)-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man-(1->6)]-alpha- D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 5A1,2)
Bound ligand (Het Group name = )
matches with 78.57% similarity
|
+
|
3
×
beta-D-mannose.CC -!- This family of mammalian enzymes, located in the Golgi system,
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:16197-16207
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanism of class 1 (glycosylhydrolase family 47) {alpha}-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control.
|
|
K.Karaveg,
A.Siriwardena,
W.Tempel,
Z.J.Liu,
J.Glushka,
B.C.Wang,
K.W.Moremen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Quality control in the endoplasmic reticulum (ER) determines the fate of newly
synthesized glycoproteins toward either correct folding or disposal by
ER-associated degradation. Initiation of the disposal process involves selective
trimming of N-glycans attached to misfolded glycoproteins by ER
alpha-mannosidase I and subsequent recognition by the ER degradation-enhancing
alpha-mannosidase-like protein family of lectins, both members of
glycosylhydrolase family 47. The unusual inverting hydrolytic mechanism
catalyzed by members of this family is investigated here by a combination of
kinetic and binding analyses of wild type and mutant forms of human ER
alpha-mannosidase I as well as by structural analysis of a co-complex with an
uncleaved thiodisaccharide substrate analog. These data reveal the roles of
potential catalytic acid and base residues and the identification of a novel
(3)S(1) sugar conformation for the bound substrate analog. The co-crystal
structure described here, in combination with the (1)C(4) conformation of a
previously identified co-complex with the glycone mimic, 1-deoxymannojirimycin,
indicates that glycoside bond cleavage proceeds through a least motion
conformational twist of a properly predisposed substrate in the -1 subsite. A
novel (3)H(4) conformation is proposed as the exploded transition state.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
FIG. 5. Normalized F[o] - F[c] disaccharide electron
density map for the thioin the active site of ERManI and
comparison of the sugar ring conformations with the enzyme-bound
conformation of dMNJ in the -1 subsite and the M7 mannose
residue in the +1 subsite. A, a stereographic representation of
the difference electron density for the omitted inhibitor in the
ERManI-thiodisaccharide cocomplex. The inhibitor model is shown
to aid in map interpretation. The reducing terminal Man-  -O-CH[3]
is shown at the top, labeled as the +1 subsite residue, and the
nonreducing terminal Man residue in the 3S[1] conformation is
labeled as the -1 subsite residue. Carbon and sulfur atoms in
the structures are labeled as a reference. The electron density
map was contoured at 3 -O-CH[3]
is shown at the top, labeled as the +1 subsite residue, and the
nonreducing terminal Man residue in the 3S[1] conformation is
labeled as the -1 subsite residue. Carbon and sulfur atoms in
the structures are labeled as a reference. The electron density
map was contoured at 3  for the gray mesh and
10 for the gray mesh and
10  for the red mesh,
demonstrating the significant electron density at the glycosidic
sulfur, the O-3' and O-4' hydroxyls of the +1 residue, and the
O-2', O-3', and O-4' hydroxyls of the -1 residue. B, the protein
structure of the ER-ManI-thiodisaccharide co-complex was aligned
with the corresponding protein structures of the
ERManI·dMNJ co-complex (20) and the co-complex of yeast
ERManI containing a Man[5]GlcNAc[2] glycan in the active site
(21) using Swiss-PdbViewer (version 3.7) (55). Displayed in the
figure are the structures of the thiodisaccharide (yellow stick
figure), dMNJ (green stick figure), and the M7 residue of the
Man[5]GlcNAc[2] glycan in the +1 subsite (white stick figure;
see Ref. 19 for oligosaccharide residue nomenclature). Carbon
and sulfur atoms in the structures are labeled as a reference.
The M7 residue is in an identical conformation as the +1 residue
of the thiodisaccharide and in a similar position except for an
offset of 0.5-0.7 Å resulting from the longer C-S bond
lengths of the thiodisaccharide. The positioning of the -1
subsite residues (dMNJ versus the -1 residue of the
thiodisaccharide) were virtually identical at the C-2, C-3, and
C-4 positions. The main differences between the two structures
were found in the equivalent of the C-1, O-5, C-5, and C-6
positions. for the red mesh,
demonstrating the significant electron density at the glycosidic
sulfur, the O-3' and O-4' hydroxyls of the +1 residue, and the
O-2', O-3', and O-4' hydroxyls of the -1 residue. B, the protein
structure of the ER-ManI-thiodisaccharide co-complex was aligned
with the corresponding protein structures of the
ERManI·dMNJ co-complex (20) and the co-complex of yeast
ERManI containing a Man[5]GlcNAc[2] glycan in the active site
(21) using Swiss-PdbViewer (version 3.7) (55). Displayed in the
figure are the structures of the thiodisaccharide (yellow stick
figure), dMNJ (green stick figure), and the M7 residue of the
Man[5]GlcNAc[2] glycan in the +1 subsite (white stick figure;
see Ref. 19 for oligosaccharide residue nomenclature). Carbon
and sulfur atoms in the structures are labeled as a reference.
The M7 residue is in an identical conformation as the +1 residue
of the thiodisaccharide and in a similar position except for an
offset of 0.5-0.7 Å resulting from the longer C-S bond
lengths of the thiodisaccharide. The positioning of the -1
subsite residues (dMNJ versus the -1 residue of the
thiodisaccharide) were virtually identical at the C-2, C-3, and
C-4 positions. The main differences between the two structures
were found in the equivalent of the C-1, O-5, C-5, and C-6
positions.
|
 |
Figure 7.
FIG. 7. Interactions between the thiodisaccharide and the
+1 and -1 binding sites in human ERManI. Shown is a schematic
diagram (left panel) of the interactions between the
thiodisaccharide and ERManI in the -1 and +1 subsites,
demonstrating hydrogen bonding interactions (green dotted
lines), direct coordination of the enzyme-associated Ca^2+ ion
(blue dotted lines), hydrophobic stacking of Phe^659 with the
C-4-C-5-C-6 region of the -1 residue (black dotted lines), and
proposed acid-catalyzed through-water (W8) protonation of the
glycosidic oxygen (sulfur in the thiodisaccharide) and the
base-catalyzed (Glu599) attack by the water nucleophile (W5)
(red dotted lines). Residue numbering of amino acid side chains
in the respective subsites is indicated. The stereo view (center
and right) illustrates a stick diagram of the interaction
between the thiodisaccharide residues in the -1 and +1 subsites
relative to the residues examined by mutagenesis here.
Coordination to the Ca^2+ ion (blue dotted lines), and the
proposed nucleophile trajectory and acid protonation of the
glycosidic oxygen (red dotted lines) are indicated. The small
red and green space fill structures representing the water
molecules and carbonyl oxygen and O-  of Thr688 that
coordinate the Ca^2+ ion are as described in the legend to Fig.
2. The green dotted lines indicate hydrogen bonds between the
respective residues. of Thr688 that
coordinate the Ca^2+ ion are as described in the legend to Fig.
2. The green dotted lines indicate hydrogen bonds between the
respective residues.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
16197-16207)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Aebi,
R.Bernasconi,
S.Clerc,
and
M.Molinari
(2010).
N-glycan structures: recognition and processing in the ER.
|
| |
Trends Biochem Sci,
35,
74-82.
|
 |
|
|
|
|
 |
M.D.Suits,
Y.Zhu,
E.J.Taylor,
J.Walton,
D.L.Zechel,
H.J.Gilbert,
and
G.J.Davies
(2010).
Structure and kinetic investigation of Streptococcus pyogenes family GH38 alpha-mannosidase.
|
| |
PLoS One,
5,
e9006.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.V.Vuong,
and
D.B.Wilson
(2010).
Glycoside hydrolases: catalytic base/nucleophile diversity.
|
| |
Biotechnol Bioeng,
107,
195-205.
|
 |
|
|
|
|
 |
Y.Zhu,
M.D.Suits,
A.J.Thompson,
S.Chavan,
Z.Dinev,
C.Dumon,
N.Smith,
K.W.Moremen,
Y.Xiang,
A.Siriwardena,
S.J.Williams,
H.J.Gilbert,
and
G.J.Davies
(2010).
Mechanistic insights into a Ca2+-dependent family of alpha-mannosidases in a human gut symbiont.
|
| |
Nat Chem Biol,
6,
125-132.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.H.Cormier,
T.Tamura,
J.C.Sunryd,
and
D.N.Hebert
(2009).
EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex.
|
| |
Mol Cell,
34,
627-633.
|
 |
|
|
|
|
 |
J.Zhou,
C.Z.Lin,
X.Z.Zheng,
X.J.Lin,
W.J.Sang,
S.H.Wang,
Z.H.Wang,
D.Ebbole,
and
G.D.Lu
(2009).
Functional analysis of an alpha-1,2-mannosidase from Magnaporthe oryzae.
|
| |
Curr Genet,
55,
485-496.
|
 |
|
|
|
|
 |
D.J.Vocadlo,
and
G.J.Davies
(2008).
Mechanistic insights into glycosidase chemistry.
|
| |
Curr Opin Chem Biol,
12,
539-555.
|
 |
|
|
|
|
 |
D.W.Abbott,
and
A.B.Boraston
(2008).
Structural biology of pectin degradation by Enterobacteriaceae.
|
| |
Microbiol Mol Biol Rev,
72,
301.
|
 |
|
|
|
|
 |
H.M.Mora-Montes,
E.López-Romero,
S.Zinker,
P.Ponce-Noyola,
and
A.Flores-Carreón
(2008).
Conversion of alpha1,2-mannosidase E-I from Candida albicans to alpha1,2-mannosidase E-II by limited proteolysis.
|
| |
Antonie Van Leeuwenhoek,
93,
61-69.
|
 |
|
|
|
|
 |
K.N.Beverly,
M.R.Sawaya,
E.Schmid,
and
C.M.Koehler
(2008).
The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23.
|
| |
J Mol Biol,
382,
1144-1156.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.N.Kirschner,
A.B.Yongye,
S.M.Tschampel,
J.González-Outeiriño,
C.R.Daniels,
B.L.Foley,
and
R.J.Woods
(2008).
GLYCAM06: A generalizable biomolecular force field. Carbohydrates.
|
| |
J Comput Chem,
29,
622-655.
|
 |
|
|
|
|
 |
T.M.Gloster,
J.P.Turkenburg,
J.R.Potts,
B.Henrissat,
and
G.J.Davies
(2008).
Divergence of catalytic mechanism within a glycosidase family provides insight into evolution of carbohydrate metabolism by human gut flora.
|
| |
Chem Biol,
15,
1058-1067.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.D.Lobsanov,
T.Yoshida,
T.Desmet,
W.Nerinckx,
P.Yip,
M.Claeyssens,
A.Herscovics,
and
P.L.Howell
(2008).
Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
227-236.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Molinari
(2007).
N-glycan structure dictates extension of protein folding or onset of disposal.
|
| |
Nat Chem Biol,
3,
313-320.
|
 |
|
|
|
|
 |
P.Marchetti,
M.Bugliani,
R.Lupi,
L.Marselli,
M.Masini,
U.Boggi,
F.Filipponi,
G.C.Weir,
D.L.Eizirik,
and
M.Cnop
(2007).
The endoplasmic reticulum in pancreatic beta cells of type 2 diabetes patients.
|
| |
Diabetologia,
50,
2486-2494.
|
 |
|
|
|
|
 |
R.L.Rich,
and
D.G.Myszka
(2006).
Survey of the year 2005 commercial optical biosensor literature.
|
| |
J Mol Recognit,
19,
478-534.
|
 |
|
|
|
|
 |
V.A.Money,
N.L.Smith,
A.Scaffidi,
R.V.Stick,
H.J.Gilbert,
and
G.J.Davies
(2006).
Substrate distortion by a lichenase highlights the different conformational itineraries harnessed by related glycoside hydrolases.
|
| |
Angew Chem Int Ed Engl,
45,
5136-5140.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Park,
L.Meng,
L.H.Stanton,
R.E.Collins,
S.W.Mast,
X.Yi,
H.Strachan,
and
K.W.Moremen
(2005).
Characterization of a human core-specific lysosomal {alpha}1,6-mannosidase involved in N-glycan catabolism.
|
| |
J Biol Chem,
280,
37204-37216.
|
 |
|
|
|
|
 |
D.N.Hebert,
S.C.Garman,
and
M.Molinari
(2005).
The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags.
|
| |
Trends Cell Biol,
15,
364-370.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

