 |
PDBsum entry 1wzf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1wzf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.18
- heme oxygenase (biliverdin-producing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
heme b + 3 reduced [NADPH--hemoprotein reductase] + 3 O2 = biliverdin IXalpha + CO + Fe2+ + 3 oxidized [NADPH--hemoprotein reductase] + 3 H2O + H+
|
 |
 |
 |
 |
 |
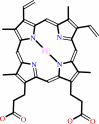 heme b
heme b
|
+
|
3
×
reduced [NADPH--hemoprotein reductase]
|
+
|
 3
×
O2
3
×
O2
|
=
|
biliverdin IXalpha
|
+
|
 CO
CO
|
+
|
Fe(2+)
|
+
|
3
×
oxidized [NADPH--hemoprotein reductase]
|
+
|
3
×
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:9416-9421
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Design of metal cofactors activated by a protein-protein electron transfer system.
|
|
T.Ueno,
N.Yokoi,
M.Unno,
T.Matsui,
Y.Tokita,
M.Yamada,
M.Ikeda-Saito,
H.Nakajima,
Y.Watanabe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protein-to-protein electron transfer (ET) is a critical process in biological
chemistry for which fundamental understanding is expected to provide a wealth of
applications in biotechnology. Investigations of protein-protein ET systems in
reductive activation of artificial cofactors introduced into proteins remains
particularly challenging because of the complexity of interactions between the
cofactor and the system contributing to ET. In this work, we construct an
artificial protein-protein ET system, using heme oxygenase (HO), which is known
to catalyze the conversion of heme to biliverdin. HO uses electrons provided
from NADPH/cytochrome P450 reductase (CPR) through protein-protein complex
formation during the enzymatic reaction. We report that a Fe(III)(Schiff-base),
in the place of the active-site heme prosthetic group of HO, can be reduced by
NADPH/CPR. The crystal structure of the Fe(10-CH(2)CH(2)COOH-Schiff-base).HO
composite indicates the presence of a hydrogen bond between the propionic acid
carboxyl group and Arg-177 of HO. Furthermore, the ET rate from NADPH/CPR to the
composite is 3.5-fold faster than that of Fe(Schiff-base).HO, although the redox
potential of Fe(10-CH(2)CH(2)COOH-Schiff-base).HO (-79 mV vs. NHE) is lower than
that of Fe(Schiff-base).HO (+15 mV vs. NHE), where NHE is normal hydrogen
electrode. This work describes a synthetic metal complex activated by means of a
protein-protein ET system, which has not previously been reported. Moreover, the
result suggests the importance of the hydrogen bond for the ET reaction of HO.
Our Fe(Schiff-base).HO composite model system may provide insights with regard
to design of ET biosystems for sensors, catalysts, and electronics devices.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Catalytic reaction of HO. (A) Enzymatic cycle of
heme metabolism to biliverdin catalyzed by HO. The reactions
shaded in blue indicate the ET step from CPR to heme to be used
for the reduction of Fe^III(Schiff-base) complexes. (B) Chemical
structures of heme (Left) and Fe^III(Schiff-base) (Right).
|
 |
Figure 2.
Fig. 2. Close-up views of coordination geometries of the
metal cofactors in the HO active-site region. Refined model
coordinates for the active site region are superimposed on 2F[o]
– F[c] maps contoured at 1.0  , except for
heme·HO, with elements of the protein and cofactors in
ball-and-stick models for top (Left) and side (Right) views.
Green is carbon, and purple, red, and pink are nitrogen, oxygen,
and iron, respectively. (A) Heme·HO. (B) 1·HO
refined to 1.35 Å. (C) 2·HO refined to 1.85
Å. (D) 3·HO refined to 1.75 Å. , except for
heme·HO, with elements of the protein and cofactors in
ball-and-stick models for top (Left) and side (Right) views.
Green is carbon, and purple, red, and pink are nitrogen, oxygen,
and iron, respectively. (A) Heme·HO. (B) 1·HO
refined to 1.35 Å. (C) 2·HO refined to 1.85
Å. (D) 3·HO refined to 1.75 Å.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Y.Lee,
A.Hille,
I.Kitanovic,
P.Jesse,
G.Henze,
S.Wölfl,
R.Gust,
and
A.Prokop
(2011).
[Fe(III)(salophene)Cl], a potent iron salophene complex overcomes multiple drug resistance in lymphoma and leukemia cells.
|
| |
Leuk Res,
35,
387-393.
|
 |
|
|
|
|
 |
A.Dalla Cort,
P.De Bernardin,
G.Forte,
and
F.Y.Mihan
(2010).
Metal-salophen-based receptors for anions.
|
| |
Chem Soc Rev,
39,
3863-3874.
|
 |
|
|
|
|
 |
L.J.Smith,
A.Kahraman,
and
J.M.Thornton
(2010).
Heme proteins--diversity in structural characteristics, function, and folding.
|
| |
Proteins,
78,
2349-2368.
|
 |
|
|
|
|
 |
C.L.Davies,
E.L.Dux,
and
A.K.Duhme-Klair
(2009).
Supramolecular interactions between functional metal complexes and proteins.
|
| |
Dalton Trans,
(),
10141-10154.
|
 |
|
|
|
|
 |
X.Zhang
(2009).
4-Chloro-2-[(E)-(2-chloro-phen-yl)imino-meth-yl]phenol.
|
| |
Acta Crystallogr Sect E Struct Rep Online,
65,
o512.
|
 |
|
|
|
|
 |
X.Zhang
(2009).
2-Bromo-4-chloro-6-[(E)-p-tolyl-imino-meth-yl]phenol.
|
| |
Acta Crystallogr Sect E Struct Rep Online,
65,
o513.
|
 |
|
|
|
|
 |
N.Yokoi,
T.Ueno,
M.Unno,
T.Matsui,
M.Ikeda-Saito,
and
Y.Watanabe
(2008).
Ligand design for the improvement of stability of metal complex.protein hybrids.
|
| |
Chem Commun (Camb),
(),
229-231.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

