 |
PDBsum entry 1vzw

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.3.1.16
- 1-(5-phosphoribosyl)-5-
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Histidine Biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-(5-phospho-beta-D-ribosyl)-5-[(5-phospho-beta-D- ribosylamino)methylideneamino]imidazole-4-carboxamide = 5-[(5-phospho-1- deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.1.24
- phosphoribosylanthranilate isomerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-(5-phospho-beta-D-ribosyl)anthranilate = 1-(2-carboxyphenylamino)-1- deoxy-D-ribulose 5-phosphate
|
 |
 |
 |
 |
 |
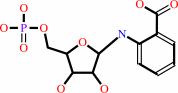 N-(5-phospho-beta-D-ribosyl)anthranilate
N-(5-phospho-beta-D-ribosyl)anthranilate
|
=
|
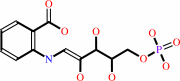 1-(2-carboxyphenylamino)-1- deoxy-D-ribulose 5-phosphate
1-(2-carboxyphenylamino)-1- deoxy-D-ribulose 5-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO Rep
6:134-139
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Two-fold repeated (betaalpha)4 half-barrels may provide a molecular tool for dual substrate specificity.
|
|
J.Kuper,
C.Doenges,
M.Wilmanns.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Some bacterial genomes contain an incomplete set of genes encoding
phosphoribosyl isomerases, raising the question of whether there exists
broadened substrate specificity for the missing gene products. To investigate
the underlying molecular principles of this hypothesis, we have determined the
crystal structure of the bifunctional enzyme PriA from Streptomyces coelicolor
at 1.8 A resolution. It consists of a (betaalpha)(8)-barrel fold that is
assembled by two symmetric (betaalpha)(4) half-barrels. The structure shows how
its active site may catalyse the isomerization reactions of two different
substrates, and we provide a plausible model of how the smaller of the two
substrates could be bound in two different orientations. Our findings expand the
half-barrel ancestor concept by demonstrating that symmetry-related half-barrels
could provide a smart solution to cope with dual substrate specificity. The data
may help to unravel molecular rationales regarding how organisms with miniature
genomes can keep central biological pathways functional.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 HisA and TrpF catalyse similar reactions in histidine
and tryptophan biosynthesis. HisA and TrpF catalyse the
isomerizations of the aminoaldoses ProFAR and PRA to the
aminoketoses
N'-((5'-phosphoribulosyl)formimino)-5-aminoimidazole-4-carboxamide
ribonucleotide (PRFAR) and
1-(o-carboxyphenylamino)-1-deoxyribulose 5-phosphate (CdRP). The
PriA protein catalyses both reactions in Streptomyces coelicolor
and Mycobacterium tuberculosis. These organisms lack a trpf gene.
|
 |
Figure 4.
Figure 4 Active site of PriA. (A) Superimposition of the active
sites of PriA (blue) and HisA (green). Active-site residues and
the two bound sulphate ions are shown in stick mode. The carbon
atoms are in the colour of the corresponding C[  ]trace.
Oxygen molecules are coloured red and sulphur in yellow. (B)
Superimposition of the active sites of PriA (blue) and TrpF
(black). Active-site residues and the two bound sulphate ions
are shown in stick mode using atom-type colours. (C) PriA active
sites with the HisA product analogue rPRFAR. The rPRFAR
coordinates have been taken from the superimposed HisF structure
(not shown) in the presence of rPRFAR (Chaudhuri et al, 2001).
Residues most probably involved in catalysis are highlighted
green, surrounded by red circles, in this panel and in all
following panels with modelled complexes. (D) PriA active site
with the TrpF product analogue rCdRP. The rCdRP coordinates were
obtained from the superimposed TrpF structure in complex with
rCdRP (Henn-Sax et al, 2002). (E) PriA active site with the TrpF
product analogue rCdRP modelled binding to the second phosphate
binding site. The rCdRP coordinates were obtained from the TrpF
structure in complex with rCdRP (Henn-Sax et al, 2002)
superimposed with the C-terminal half-barrel of PriA. Complete
PriA was then superimposed using the coordinates obtained from
the C-terminal half-barrel. (A -E) Prepared with DINO
(http://www.dino3d.org) and rendered with POVRAY 3.6
(http://www.povray.org). ]trace.
Oxygen molecules are coloured red and sulphur in yellow. (B)
Superimposition of the active sites of PriA (blue) and TrpF
(black). Active-site residues and the two bound sulphate ions
are shown in stick mode using atom-type colours. (C) PriA active
sites with the HisA product analogue rPRFAR. The rPRFAR
coordinates have been taken from the superimposed HisF structure
(not shown) in the presence of rPRFAR (Chaudhuri et al, 2001).
Residues most probably involved in catalysis are highlighted
green, surrounded by red circles, in this panel and in all
following panels with modelled complexes. (D) PriA active site
with the TrpF product analogue rCdRP. The rCdRP coordinates were
obtained from the superimposed TrpF structure in complex with
rCdRP (Henn-Sax et al, 2002). (E) PriA active site with the TrpF
product analogue rCdRP modelled binding to the second phosphate
binding site. The rCdRP coordinates were obtained from the TrpF
structure in complex with rCdRP (Henn-Sax et al, 2002)
superimposed with the C-terminal half-barrel of PriA. Complete
PriA was then superimposed using the coordinates obtained from
the C-terminal half-barrel. (A -E) Prepared with DINO
(http://www.dino3d.org) and rendered with POVRAY 3.6
(http://www.povray.org).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO Rep
(2005,
6,
134-139)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.V.Due,
J.Kuper,
A.Geerlof,
J.P.Kries,
and
M.Wilmanns
(2011).
Bisubstrate specificity in histidine/tryptophan biosynthesis isomerase from Mycobacterium tuberculosis by active site metamorphosis.
|
| |
Proc Natl Acad Sci U S A,
108,
3554-3559.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Kim,
J.Basner,
and
B.Lee
(2010).
Detecting internally symmetric protein structures.
|
| |
BMC Bioinformatics,
11,
303.
|
 |
|
|
|
|
 |
L.Noda-García,
A.R.Camacho-Zarco,
K.Verdel-Aranda,
H.Wright,
X.Soberón,
V.Fülöp,
and
F.Barona-Gómez
(2010).
Identification and analysis of residues contained on beta --> alpha loops of the dual-substrate (beta alpha)8 phosphoribosyl isomerase A specific for its phosphoribosyl anthranilate isomerase activity.
|
| |
Protein Sci,
19,
535-543.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Carbonell,
and
J.L.Faulon
(2010).
Molecular signatures-based prediction of enzyme promiscuity.
|
| |
Bioinformatics,
26,
2012-2019.
|
 |
|
|
|
|
 |
J.Claren,
C.Malisi,
B.Höcker,
and
R.Sterner
(2009).
Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha)8-barrel protein scaffolds.
|
| |
Proc Natl Acad Sci U S A,
106,
3704-3709.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Payandeh,
and
E.F.Pai
(2007).
Enzyme-driven speciation: crystallizing Archaea via lipid capture.
|
| |
J Mol Evol,
64,
364-374.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

