 |
PDBsum entry 1vz7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.35
- glutamate N-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N2-acetyl-L-ornithine + L-glutamate = N-acetyl-L-glutamate + L-ornithine
|
 |
 |
 |
 |
 |
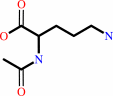 N(2)-acetyl-L-ornithine
N(2)-acetyl-L-ornithine
|
+
|
 L-glutamate
L-glutamate
|
=
|
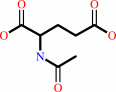 N-acetyl-L-glutamate
N-acetyl-L-glutamate
|
+
|
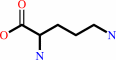 L-ornithine
L-ornithine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem J
385:565-573
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray crystal structure of ornithine acetyltransferase from the clavulanic acid biosynthesis gene cluster.
|
|
J.M.Elkins,
N.J.Kershaw,
C.J.Schofield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The orf6 gene from the clavulanic acid biosynthesis gene cluster encodes an OAT
(ornithine acetyltransferase). Similar to other OATs the enzyme has been shown
to catalyse the reversible transfer of an acetyl group from N-acetylornithine to
glutamate. OATs are Ntn (N-terminal nucleophile) enzymes, but are distinct from
the better-characterized Ntn hydrolase enzymes as they catalyse acetyl transfer
rather than a hydrolysis reaction. In the present study, we describe the X-ray
crystal structure of the OAT, corresponding to the orf6 gene product, to 2.8 A
(1 A=0.1 nm) resolution. The larger domain of the structure consists of an
alphabetabetaalpha sandwich as in the structures of Ntn hydrolase enzymes.
However, differences in the connectivity reveal that OATs belong to a structural
family different from that of other structurally characterized Ntn enzymes, with
one exception: unexpectedly, the alphabetabetaalpha sandwich of ORF6 (where ORF
stands for open reading frame) displays the same fold as an DmpA
(L-aminopeptidase D-ala-esterase/amidase from Ochrobactrum anthropi), and so the
OATs and DmpA form a new structural subfamily of Ntn enzymes. The structure
reveals an alpha2beta2-heterotetrameric oligomerization state in which the
intermolecular interface partly defines the active site. Models of the
enzyme-substrate complexes suggest a probable oxyanion stabilization mechanism
as well as providing insight into how the enzyme binds its two differently
charged substrates.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Sankaranarayanan,
C.R.Garen,
M.M.Cherney,
M.Yuan,
C.Lee,
and
M.N.James
(2009).
Preliminary X-ray crystallographic analysis of ornithine acetyltransferase (Rv1653) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
173-176.
|
 |
|
|
|
|
 |
B.Geueke,
and
H.P.Kohler
(2007).
Bacterial beta-peptidyl aminopeptidases: on the hydrolytic degradation of beta-peptides.
|
| |
Appl Microbiol Biotechnol,
74,
1197-1204.
|
 |
|
|
|
|
 |
Y.Xu,
B.Labedan,
and
N.Glansdorff
(2007).
Surprising arginine biosynthesis: a reappraisal of the enzymology and evolution of the pathway in microorganisms.
|
| |
Microbiol Mol Biol Rev,
71,
36-47.
|
 |
|
|
|
|
 |
D.Maes,
M.Crabeel,
C.Van de Weerdt,
J.Martial,
E.Peeters,
D.Charlier,
K.Decanniere,
C.Vanhee,
L.Wyns,
and
I.Zegers
(2006).
Crystallization of ornithine acetyltransferase from yeast by counter-diffusion and preliminary X-ray study.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1294-1297.
|
 |
|
|
|
|
 |
H.Arulanantham,
N.J.Kershaw,
K.S.Hewitson,
C.E.Hughes,
J.E.Thirkettle,
and
C.J.Schofield
(2006).
ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid.
|
| |
J Biol Chem,
281,
279-287.
|
 |
|
|
|
|
 |
H.Cheng,
and
N.V.Grishin
(2005).
DOM-fold: a structure with crossing loops found in DmpA, ornithine acetyltransferase, and molybdenum cofactor-binding domain.
|
| |
Protein Sci,
14,
1902-1910.
|
 |
|
|
|
|
 |
K.Michalska,
K.Brzezinski,
and
M.Jaskolski
(2005).
Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate.
|
| |
J Biol Chem,
280,
28484-28491.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

